Genetic Variability and Population Structure of the Potential Bioenergy Crop Miscanthus sinensis (Poaceae) in Southwest China Based on SRAP Markers
Abstract
:1. Introduction
2. Results and Discussion
2.1. Polymorphism of SRAP Markers
2.2. Genetic Diversity and AMOVA Analysis
| Pirmer Pairs | Sequence 5'→3' | Total Number of Bands | Number of Polymorphic Bands | Percentage of Polymorphic Bands (%) | Polymorphic Information Content (PIC) |
|---|---|---|---|---|---|
| Me1 + em3 | TGAGTCCAAACCGGATA GACTGCGTACGAATTGAC | 15 | 15 | 100.00 | 0.33 |
| Me1 + em8 | TGAGTCCAAACCGGATA GACTGCGTACGAATTCTG | 16 | 13 | 81.25 | 0.34 |
| Me1 + em10 | TGAGTCCAAACCGGATA GACTGCGTACGAATTCAG | 21 | 21 | 100.00 | 0.38 |
| Me2 + em1 | GACTGCGTACGAATTTGC GACTGCGTACGAATTAAT | 21 | 21 | 100.00 | 0.39 |
| Me2 + em9 | GACTGCGTACGAATTTGC GACTGCGTACGAATTCGA | 17 | 14 | 82.35 | 0.28 |
| Me2 + em10 | GACTGCGTACGAATTTGC GACTGCGTACGAATTCAG | 29 | 28 | 96.55 | 0.40 |
| Me3 + em5 | TGAGTCCAAACCGGAAT GACTGCGTACGAATTACC | 18 | 15 | 83.33 | 0.23 |
| Me3 + em9 | TGAGTCCAAACCGGAAT GACTGCGTACGAATTCGA | 18 | 17 | 94.44 | 0.31 |
| Me3 + em10 | TGAGTCCAAACCGGAAT GACTGCGTACGAATTCAG | 24 | 24 | 100.00 | 0.34 |
| Me4 + em1 | TGAGTCCAAACCGGACC GACTGCGTACGAATTAAT | 16 | 16 | 100.00 | 0.28 |
| Me4 + em7 | TGAGTCCAAACCGGACC GACTGCGTACGAATTCAA | 22 | 21 | 95.45 | 0.35 |
| Me4 + em9 | TGAGTCCAAACCGGACC GACTGCGTACGAATTCGA | 20 | 18 | 90.00 | 0.29 |
| Me4 + em10 | TGAGTCCAAACCGGACC GACTGCGTACGAATTCAG | 20 | 19 | 95.00 | 0.41 |
| Me5 + em2 | TGAGTCCAAACCGGAAG GACTGCGTACGAATTTGC | 21 | 19 | 90.48 | 0.30 |
| Me5 + em4 | TGAGTCCAAACCGGAAG GACTGCGTACGAATTTGA | 19 | 17 | 89.47 | 0.31 |
| Me5 + em8 | TGAGTCCAAACCGGAAG GACTGCGTACGAATTCTG | 25 | 25 | 100.00 | 0.38 |
| Me5 + em10 | TGAGTCCAAACCGGAAG GACTGCGTACGAATTCAG | 23 | 23 | 100.00 | 0.33 |
| Me6 + em7 | TGAGTCCAAACCGGTAA GACTGCGTACGAATTCAA | 18 | 17 | 94.44 | 0.34 |
| Me6 + em8 | TGAGTCCAAACCGGTAA GACTGCGTACGAATTCTG | 27 | 23 | 85.19 | 0.25 |
| Me6 + em10 | TGAGTCCAAACCGGTAA GACTGCGTACGAATTCAG | 30 | 30 | 100.00 | 0.38 |
| Me7 + em1 | TGAGTCCAAACCGGTCC GACTGCGTACGAATTAAT | 13 | 9 | 69.23 | 0.28 |
| Me7 + em5 | TGAGTCCAAACCGGTCC GACTGCGTACGAATTACC | 24 | 22 | 91.67 | 0.33 |
| Me7 + em8 | TGAGTCCAAACCGGTCC GACTGCGTACGAATTCTG | 16 | 15 | 93.75 | 0.33 |
| Me7 + em10 | TGAGTCCAAACCGGTCC GACTGCGTACGAATTCAG | 24 | 23 | 95.83 | 0.39 |
| Me8 + em5 | TGAGTCCAAACCGGTGC GACTGCGTACGAATTACC | 22 | 22 | 100.00 | 0.37 |
| Me8 + em7 | TGAGTCCAAACCGGTGC GACTGCGTACGAATTCAA | 19 | 18 | 94.74 | 0.40 |
| Me8 + em9 | TGAGTCCAAACCGGTGC GACTGCGTACGAATTCGA | 23 | 23 | 100.00 | 0.32 |
| Me9 + em1 | TGAGTCCAAACCGGTAG GACTGCGTACGAATTAAT | 25 | 24 | 96.00 | 0.33 |
| Me9 + em5 | TGAGTCCAAACCGGTAG GACTGCGTACGAATTACC | 18 | 16 | 88.89 | 0.36 |
| Me9 + em8 | TGAGTCCAAACCGGTAG GACTGCGTACGAATTCTG | 17 | 15 | 88.24 | 0.34 |
| Me9 + em9 | TGAGTCCAAACCGGTAG GACTGCGTACGAATTCGA | 21 | 19 | 90.48 | 0.36 |
| Me10 + em1 | TGAGTCCAAACCGGTTG GACTGCGTACGAATTAAT | 22 | 21 | 95.45 | 0.38 |
| Me10 + em2 | TGAGTCCAAACCGGTTG GACTGCGTACGAATTTGC | 24 | 23 | 95.83 | 0.38 |
| Total | 688 | 646 | |||
| Mean | 20.85 | 19.58 | 93.27 | 0.34 |
| Population Identity | Sample Size | H | I | P |
|---|---|---|---|---|
| Pop1 | 15 | 0.2712 | 0.4062 | 77.91% |
| Pop2 | 13 | 0.2939 | 0.4351 | 79.80% |
| Pop3 | 12 | 0.2877 | 0.4261 | 77.76% |
| Pop4 | 13 | 0.2852 | 0.4237 | 78.78% |
| Pop5 | 11 | 0.2727 | 0.4042 | 73.98% |
| Pop6 | 8 | 0.2599 | 0.3830 | 68.46% |
| Pop7 | 11 | 0.2849 | 0.4206 | 76.60% |
| Pop8 | 10 | 0.2792 | 0.4127 | 74.71% |
| Pop9 | 8 | 0.2734 | 0.4035 | 73.11% |
| Pop10 | 12 | 0.2799 | 0.4130 | 75.00% |
| Pop11 | 9 | 0.2580 | 0.3819 | 70.06% |
| Pop12 | 5 | 0.0787 | 0.1162 | 21.22% |
| Pop13 | 15 | 0.2773 | 0.4122 | 77.91% |
| Pop14 | 5 | 0.2480 | 0.3624 | 62.79% |
| Pop15 | 18 | 0.3052 | 0.4540 | 84.30% |
| Pop16 | 14 | 0.2828 | 0.4209 | 79.22% |
| Pop17 | 16 | 0.2591 | 0.3910 | 77.91% |
| Pop18 | 11 | 0.2716 | 0.4052 | 76.45% |
| Pop19 | 8 | 0.2679 | 0.3930 | 69.04% |
| Pop20 | 15 | 0.2731 | 0.4090 | 78.49% |
| Pop21 | 12 | 0.2708 | 0.4024 | 75.00% |
| Pop22 | 6 | 0.2360 | 0.3484 | 62.65% |
| Pop23 | 6 | 0.2114 | 0.3116 | 55.81% |
| Pop24 | 7 | 0.1847 | 0.2700 | 47.24% |
| Mean | 0.2589 | 0.3836 | 70.59% | |
| Within Species | 260 | 0.3377 | 0.5032 | 93.90% |
| Source | Degree of Freedom | Sum of Squares | Summary of Matches | Percentage of Variation |
|---|---|---|---|---|
| Among Pops | 23 | 6468.464 | 281.238 | 14% |
| Within Pops | 236 | 24162.059 | 102.382 | 86% |
| Total | 259 | 30630.523 | 100% |
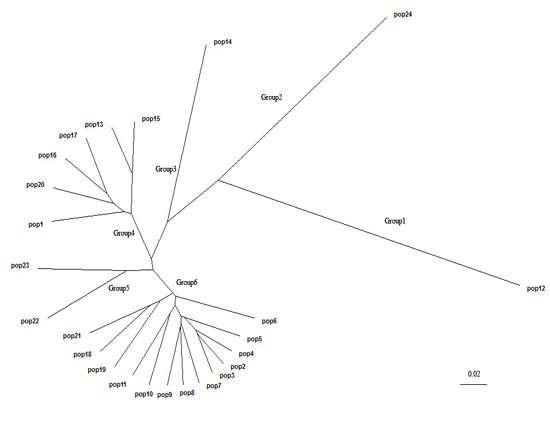
2.3. Population Structure and Cluster Analysis


3. Experimental Section
3.1. Plant Material Collection
| Population Identity | Sample Size | Region | Altitude (m) | Latitude (N) | Longitude (E) | Habital |
|---|---|---|---|---|---|---|
| Pop1 | 15 | Ya’an | 677 | 29°58'39.3'' | 102°59'24.5'' | Glade at hillside |
| Pop2 | 13 | Bi Feng Xia | 989 | 30°07'25.5'' | 102°59'48.6'' | Highway side |
| Pop3 | 12 | Bao Xing | 1253 | 30°16'05.7'' | 102°49'19.5'' | River beach |
| Pop4 | 13 | Erlang Mountain | 2091 | 29°51'19.8'' | 102°18'58.3'' | Glade at hillside |
| Pop5 | 11 | Tuowu Mountain | 1630 | 28°59'52.9'' | 102°18'12.9'' | Glade at hillside |
| Pop6 | 8 | Niba Mountain | 1636 | 29°39'46.1'' | 102°36'25.0'' | Glade at hillside |
| Pop7 | 11 | Renshou | 471 | 30°01'05.9'' | 103°58'21.8'' | Hillside |
| Pop8 | 10 | Hongya | 493 | 29°51'22.6'' | 103°14'03.5'' | Dam |
| Pop9 | 8 | Zizhong | 350 | 29°48'50.4'' | 104°42'28.6'' | Glade in orangery |
| Pop10 | 12 | Luzhou | 241 | 28°51'01.9'' | 105°18'19.9'' | Rice field ridge |
| Pop11 | 9 | Yibin | 317 | 28°45'15.4'' | 104°36'48.5'' | Shrub at riverside |
| Pop12 | 5 | Zigong | 353 | 29°24'21.9'' | 104°49'01.4'' | Grassland |
| Pop13 | 15 | Jiangyou | 687 | 31°58'06.8'' | 105°04'38.4'' | Highway slope |
| Pop14 | 5 | Jian’ge | 611 | 32°13'25.0'' | 105°35'17.9'' | Shrub at hillside |
| Pop15 | 18 | Guangyuan | 668 | 32°39'56.5'' | 105°56'21.4'' | Shrub at hillside |
| Pop16 | 14 | Daying | 327 | 30°36'32.3'' | 105°13'59.6'' | Shrub |
| Pop17 | 16 | Banan | 476 | 29°25'20.6'' | 106°34'37.5'' | Forest edge at hillside |
| Pop18 | 11 | Nanchuan | 579 | 29°10'09.3'' | 107°06'44.1'' | Shrub at hillside |
| Pop19 | 8 | Dabai | 455 | 28°32'51.8'' | 106°51'09.6'' | Highway slope |
| Pop20 | 15 | Zunyi | 914 | 27°59'11.5'' | 106°52'25.1'' | Coniferous edge |
| Pop21 | 12 | Guiyang | 1268 | 26°28'57.2'' | 106°27'35.8'' | Bare rock |
| Pop22 | 6 | Zhenning | 1284 | 26°04'49.9'' | 105°46'55.9'' | Glade |
| Pop23 | 6 | Huangguoshu | 946 | 25°58'43.6'' | 105°39'47.1'' | Forest edge |
| Pop24 | 7 | Yuxi | 1721 | 24°11'53.2'' | 102°28'29.9'' | Hillside |
| Total number of individuals | 260 |
3.2. DNA Extraction
3.3. Primer Selection and PCR-SRAP Amplification

3.4. Data Analysis
4. Conclusions
Supplementary Materials
Supplementary File
Supplementary File 1Acknowledgments
Author Contributions
Conflicts of Interest
References
- John, C.C.B.; Lewandowski, I.; Andersson, B.; Basch, G.; Christian, D.G.; Kjeldsen, J.B.; Jorgensen, U.; Mortensen, J.V.; Riche, A.B.; Sshwarz, K.U.; et al. Performance of 15 Miscanthus genotypes at five sites in Europe. Agron. J. 2001, 93, 1013–1019. [Google Scholar] [CrossRef]
- Lewandowski, I.; John, C.C.B.; Scurlock, J.M.O.; Huisman, W. Miscanthus: European experience with a novel energy crop. Biomass Bioenergy 2000, 19, 209–277. [Google Scholar] [CrossRef]
- Heaton, E.A.; Long, S.P.; Voigt, T.B.; Jones, M.B.; John, C.B. Miscanthus for renewable energy generation: European Union experience and projections for Illinois. Mitig. Adapt. Strateg. Glob. Chang. 2004, 9, 433–451. [Google Scholar] [CrossRef]
- John, C.C.B.; Lewandowski, I.; Bangerth, F.; Jones, M.B. Comparative responses to water stress in stay-green, rapid and slow senescing genotypes of the biomass crop, Miscanthu. New Phytol. 2002, 154, 335–345. [Google Scholar] [CrossRef]
- Chen, S.L.; Renvoize, S.A. Miscanthus. In Flora of China; Science Press: Beijing, China, 2006; pp. 581–583. [Google Scholar]
- Jiang, J.X.; Wang, Z.H.; Tang, B.R. Development of novel chloroplast microsatellite markers for Miscanthus species (Poaceae). Am. J. Bot. 2012, 99, e230–e233. [Google Scholar] [CrossRef]
- Hodkinson, T.R.; Chase, M.W.; Lledó, M.D.; Salamin, N.; Renvoize, S.A. Phylogenetics of Miscanthus, Saccharum and related genera (Saccharinae, Andropogoneae, Poaceae) based on DNA sequences fromITS nuclear ribosomal DNA and plastid trnL intron and trnL-F intergenic spacers. J. Plant Res. 2002, 115, 381–392. [Google Scholar] [CrossRef]
- Swaminathan, K.; Alabady, M.S.; Varala, K.; Paoli, D.E.; Ho, I.; Rokhsar, D.S.; Arumuganathan, A.K.; Ming, R.; Green, P.J.; Meyers, B.C.; et al. Genomic and small RNA sequencing of Miscanthus × giganteus shows the utility ofsorghum as a reference genome sequence forAndropogoneae grasses. Genome Biol. 2010, 11, R12. [Google Scholar]
- Greef, J.M.; Deuter, M.; Jung, C. Genetic diversity of European Miscanthus species revealed by AFLP fingerprinting. Genet. Resour. Crop Evol. 1997, 44, 185–195. [Google Scholar] [CrossRef]
- Hodkinson, T.R.; Chase, M.W.; Takahashi, C.; Leitch, I.J.; Bennett, M.D.; Renvoize, S.A. The use of DNA sequencing (ITS and trnL-F), AFLP and fluorescent in situ hybridisation to study allopolyploid Miscanthus (Poaceae). Am. J. Bot. 2002, 89, 279–286. [Google Scholar] [CrossRef]
- Lewandowski, I.; Scurtock, J.M.O.; Lindvall, E.; Christou, M. The development and current status of perennial rhizomatous grasses as energy crops in the US and Europe. Biomass Bioenergy 2003, 25, 335–361. [Google Scholar] [CrossRef]
- Xu, W.Z.; Zhang, X.Q.; Huang, L.K.; Nie, G.; Wang, J.P. Higher genetic diversity and gene flow in wild populations of Miscanthus sinensis in southwest China. Biochem. Syst. Ecol. 2013, 48, 174–181. [Google Scholar] [CrossRef]
- Huang, L.K.; Bughrara, S.S.; Zhang, X.Q. Genetic diversity of switchgrass and its relative species in Panicum genus using molecular markers. Biochem. Syst. Ecol. 2011, 39, 685–693. [Google Scholar] [CrossRef]
- Huang, L.K.; Zhang, X.Q.; Xie, W.G. Molecular diversity and population structure of the forage grass Hemarthria compressa (Poaceae) in south China based on SRAP markers. Genet. Mol. Res. 2012, 11, 2441–2450. [Google Scholar] [CrossRef]
- Cai, Q.; Fan, Y.H.; Aitken, K.; Piperidis, G.; McIntyre, C.L.; Jackson, P. Assessment of the phylogentic relationships within the Saccharum complex using AFLP markers. Acta Agron. Sin. 2005, 31, 551–559. [Google Scholar]
- Hiroyoshi, I.; Takashi, K.; Yoshihiko, T. Genetic structure of Miscanthus sinensis ssp. condensates (Poaceae) on Mmiyake Island: Implications for revegetation of volcanically devastated sites. Ecol. Res. 2005, 20, 233–238. [Google Scholar] [CrossRef]
- Hodkinson, R.T.; Chase, W.M.; Renvoize, A.S. Characterization of a genetic resource collection for Miscanthus (Saccharinae, Andropogoneae, Poaceae) using AFLP and ISSR PCR. Ann. Bot. 2002, 89, 627–636. [Google Scholar] [CrossRef]
- Zhao, H.; Wang, B.; He, J.; Yang, J.; Pan, L.; Sun, D.F.; Peng, J.H. Genetic diversity and population structure of Miscanthus sinensis germplasm in China. PLoS One 2013, 8, e75672. [Google Scholar] [CrossRef]
- Huang, Y.; Li, F.; Chen, K.S. Analysis of diversity and relationships among Chinese orchid cultivars using EST-SSR markers. Biochem. Syst. Ecol. 2010, 38, 93–102. [Google Scholar] [CrossRef]
- Li, G.; Quiros, C.F. Sequence related amplified polymorphism (SRAP), a new marker system based on a simple PCR reaction: Its application to mapping and gene tagging in Brassica. Theor. Appl. Genet. 2011, 103, 455–461. [Google Scholar]
- Peng, S.; Feng, N.; Guo, M.; Chen, Y.; Guo, Q. Genetic variation of Carthamus tinctorius L. and related species revealed by SRAP analysis. Biochem. Syst. Ecol. 2008, 36, 531–538. [Google Scholar] [CrossRef]
- Ferriol, M.; Pico, B.; Nuez, F. Genetic diversity of a germplasm collection of Cucurbita pepo using SRAP and AFLP markers. Theor. Appl. Genet. 2003, 107, 271–282. [Google Scholar] [CrossRef]
- Budak, H.; Shearman, R.C.; Parmaksiz, I.; Dweikat, I. Comparative analysis of seeded and vegetative biotype buffalograsses based on phylogenetic relationship using ISSRs, SSRs, RAPDs and SRAPs. Theor. Appl. Genet. 2004, 109, 280–288. [Google Scholar]
- Comlekcioglu, N.; Simsek, O.; Boncuk, M.; Aka-Kacar, Y. Genetic characterization of heat tolerant tomato (Solanum lycopersicon) genotypes by SRAP and RAPD markers. Genet. Mol. Res. 2010, 9, 2263–2274. [Google Scholar] [CrossRef]
- Lin, Z.X.; Zhang, X.L.; Nie, Y.C. Construction of a genetic linkage map for cotton based on SRAP. Chin. Sci. Bull. 2003, 48, 2063–2068. [Google Scholar] [CrossRef]
- Suenaga, K.; Khairallah, M.; William, H.M.; Hoisington, D.A. A new intervarietal linkage map and its application for quantitative trait locus analysis of “gigas” features in bread wheat. Genome 2005, 48, 65–75. [Google Scholar]
- Zhong, Z.L.; Jiang, J.X.; Yang, L.; Yi, Z.L. Genetic diversity analysis of Miscanthus species by maize SSR primers. Prog. Mod. Biomed. 2009, 9, 2076–2079. [Google Scholar]
- Yang, C.D.; Zhang, X.; An, Y.H.; Yuan, L.Y. Preliminary survey on distribution characteristics and population density of Miscanthus sacchariflorus community in Hubei province. Soil Land Water Conserv. China 2009, 11, 19–20. [Google Scholar]
- Zhao, H.; Yu, J.Y.; Frank, M.Y.; Luo, M.C.; Peng, J.H. Transferability of microsatellite markers from Brachypodium distachyon to Miscanthus sinensis. J. Integr. Plant Biol. 2011, 53, 232–245. [Google Scholar] [CrossRef]
- Mittermeier, R.A.; Myers, N.; Mittermeier, C.G. Hotspots: Earth’s Biologically Richest and Most Endangered Terrestrial Ecoregions; Conservation International: Washington, DC, USA, 2000; pp. 1–432. [Google Scholar]
- Nei, M. Analysis of gene diversity in subdivided population. Proc. Natl. Acad. Sci. USA 1988, 70, 3321–3323. [Google Scholar] [CrossRef]
- Clark, L.V.; Brummer, J.E.; Hall, M.; Long, S.; Peng, J.H.; Yamada, T.; Yoo, J.H.; Yu, C.Y.; Zhao, H.; Sacks, E.J. A footprint of past climate change on the diversity and population structure of Miscanthus sinensis. Ann. Bot. 2014, 114, 97–107. [Google Scholar] [CrossRef]
- Liu, Z.L.; Li, S.; Yan, G.Q.; Song, Y.; Zhao, G.F. Genetic structure and intraspecific genetic polymorphisms in natural populations of Psathyrostachys huashanica. Acta Genet. Sin. 2001, 28, 769–777. [Google Scholar]
- Zhao, L.F.; Li, S.; Pan, Y.; Yang, G.Q.; Zhao, G.F. Population differentiation of Psathyrostachys huashanica along an altitudinal gradient detected by random amplified polymorphic DNA. Acta Bot. Boreali-Occidentalia Sin. 2001, 21, 391–400. [Google Scholar]
- Hang, Y.; Jin, Y.; Lu, B.R. Genetic diversity of the endangered species Psathyrostachys huashanica in China and its strategic conservation. J. Fudan Univ. (Nat. Sci.) 2004, 43, 260–266. [Google Scholar]
- Wang, L.; Yang, J.; Guo, J.; Zhao, G.F. Genetic structure and differentiation of Psathyrostachys huashanica populations detected with RAPD markers. Front. Biol. China 2005, 25, 719–726. [Google Scholar]
- Wang, L.; Guo, J.; Zhao, G.F. Genetic diversity of the endangered and endemic species Psathyrostachys huashanica natural populations using simple sequence repeats (SSRs) markers. Biochem. Syst. Ecol. 2006, 34, 310–318. [Google Scholar] [CrossRef]
- Ma, X.; Chen, S.Y.; Zhang, X.Q.; Bai, S.Q.; Zhang, C.B. Assessment of worldwide genetic diversity of Siberian wildrye (Elymus sibiricus L.) germplasm based on gliadin analysis. Molecules 2012, 17, 4424–4434. [Google Scholar] [CrossRef]
- Hamrick, J.L.; Godt, M.J.W. Allozyme diversity in plant species. In Plant Population Genetics, Breeding and Genetic Resources; Browm, A.H.D., Clegg, M.T., Kaher, A.L., Weir, B.S., Eds.; Sinauer Associates Inc.: Sunderland, MA, USA, 1989; pp. 43–63. [Google Scholar]
- Liu, W.X.; Liu, W.H.; Wu, J. Analysis of genetic diversity in natural populations of Psathyrostachys huashanica keng using microsatellite (SSR) markers. Agric. Sci. China 2010, 9, 463–471. [Google Scholar] [CrossRef]
- Wang, Z.R. Plant Allozyme Analysis; Science Press: Beijing, China, 1998. [Google Scholar]
- Arnold, M.L. Anderson’s paradigm: Louisiana Irises and the study of evolutionary phenomena. Mol. Ecol. 2000, 9, 1687–1698. [Google Scholar] [CrossRef]
- Nei, M. Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 1978, 89, 583–590. [Google Scholar]
- Lande, R. Genetics and demography in biological conservation. Science 1988, 241, 1455–1460. [Google Scholar]
- Mills, L.S.; Smouse, P.E. Demographic consequences of inbreeding in remnant populations. Am. Nat. 1994, 144, 412–431. [Google Scholar]
- Frankham, R. Relationship of genetic variation to population size in wildlife. Conserv. Biol. 1996, 10, 1500–1508. [Google Scholar]
- Newman, D.; Pilson, D. Increased probability of extinction due to decreased genetic effective population size: Experimental populations of Clarkia pulchella. Evolution 1997, 51, 354–362. [Google Scholar] [CrossRef]
- Saccheri, I.; Kuussaari, M.; Kankare, M.; Vikman, P.; Fortelius, W.; Hanski, I. Inbreeding and extinction in a butterfly metapopulation. Nature 1998, 392, 491–494. [Google Scholar] [CrossRef]
- Hedrick, P.W. Genetics of Populations, 2nd ed.; Jones and Bartlett: Sudbury, MA, USA, 2000. [Google Scholar]
- Zietkiewicz, E.; Rafalski, A.; Labuda, D. Genome fingerprinting by simple sequence repeat (SSR) anchored polymerase chain reaction amplification. Genomics 1994, 20, 176–183. [Google Scholar] [CrossRef]
- Huang, L.K.; Zhang, X.Q.; Ma, X. Genetic differentiation among Hemarthria compressa populations in south China and its genetic relationship with H. japonica. Hereditas 2008, 145, 84–91. [Google Scholar] [CrossRef]
- Loarce, Y.; Gallego, R.; Ferrer, E. A comparative analysis of genetic relationships between rye cultivars using RFLP and RAPD markers. Euphytic 1993, 88, 107–115. [Google Scholar] [CrossRef]
- Roldán-Ruiz, I.; Dendauw, J.; van Bockstaele, E.; Depicker, A.; de Loose, M. AFLP markers reveal high polymorphic rates in ryegrasses (Lolium spp.). Mol. Breed. 2000, 6, 125–134. [Google Scholar] [CrossRef]
- Yeh, F.C.; Yang, R.C.; Boyle, T. POPGENE VERSION 1.31. Microsoft Windows-based Freeware for Population Genetic Analysis. Quick User Guide. Available online: http://www.ualberta.ca/~fyeh/popgene.pdf (accessed on 4 March 2014).
- Slatkin, M.; Barton, N.H. A comparison of three indirect methods for estimating average levels of gene flow. Evolution 1989, 43, 1349–1368. [Google Scholar] [CrossRef]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure from multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software STRUCTURE: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Rohlf, F.J. “NTSYS-pc 2.1”. Numerical Taxonomy and Multivariate Analysis System; Applied Biostatistics Inc.: New York, NY, USA, 1997. [Google Scholar]
- Peakall, R.; Smouse, P.E. GENALEX 6: Genetic analysis in Excel. Population genetic software for teaching and research. Mol. Ecol. Notes 2006, 6, 288–295. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are available from the authors.
© 2014 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Nie, G.; Zhang, X.-Q.; Huang, L.-K.; Xu, W.-Z.; Wang, J.-P.; Zhang, Y.-W.; Ma, X.; Yan, Y.-H.; Yan, H.-D. Genetic Variability and Population Structure of the Potential Bioenergy Crop Miscanthus sinensis (Poaceae) in Southwest China Based on SRAP Markers. Molecules 2014, 19, 12881-12897. https://doi.org/10.3390/molecules190812881
Nie G, Zhang X-Q, Huang L-K, Xu W-Z, Wang J-P, Zhang Y-W, Ma X, Yan Y-H, Yan H-D. Genetic Variability and Population Structure of the Potential Bioenergy Crop Miscanthus sinensis (Poaceae) in Southwest China Based on SRAP Markers. Molecules. 2014; 19(8):12881-12897. https://doi.org/10.3390/molecules190812881
Chicago/Turabian StyleNie, Gang, Xin-Quan Zhang, Lin-Kai Huang, Wen-Zhi Xu, Jian-Ping Wang, Yun-Wei Zhang, Xiao Ma, Yan-Hong Yan, and Hai-Dong Yan. 2014. "Genetic Variability and Population Structure of the Potential Bioenergy Crop Miscanthus sinensis (Poaceae) in Southwest China Based on SRAP Markers" Molecules 19, no. 8: 12881-12897. https://doi.org/10.3390/molecules190812881
APA StyleNie, G., Zhang, X. -Q., Huang, L. -K., Xu, W. -Z., Wang, J. -P., Zhang, Y. -W., Ma, X., Yan, Y. -H., & Yan, H. -D. (2014). Genetic Variability and Population Structure of the Potential Bioenergy Crop Miscanthus sinensis (Poaceae) in Southwest China Based on SRAP Markers. Molecules, 19(8), 12881-12897. https://doi.org/10.3390/molecules190812881