Accumulation of GD1α Ganglioside in MDA-MB-231 Breast Cancer Cells Expressing ST6GalNAc V
Abstract
:1. Introduction
2. Results and Discussion
2.1. Quantitative Real-Time-PCR (qPCR) Analysis of ST6GalNAc V Expression in Transfected MDA-MB-231 Cells


2.2. Flow Cytometry Analysis of α2,6-Sialylation Using Sambucus Nigra Agglutinin (SNA)

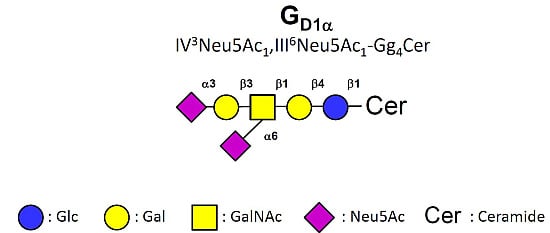
2.3. MS Analysis of GSL in ST6GalNAc V Transfected MDA-MB-231 Cells


3. Experimental Section
3.1. Cell Culture and Transfection
3.2. QPCR Analysis of ST6GalNAc V
3.3. Flow Cytometry Analysis
3.4. Extraction and Preparation of Glycolipids
3.5. Mass Spectrometry Analysis of GSL
4. Conclusions
Acknowledgments
Author Contributions
Abbreviations
| BAS | Bovine Serum Albumin |
| Cer | ceramide |
| DMEM | Dulbecco’s Modified Eagle’s Medium |
| FBS | Fetal Bovine Serum |
| GFP | green fluorescent protein |
| GSL | glycosphingolipid |
| HPRT | Hypoxanthine PhosphoRibosylTransferase |
| LacCer | Lactosylceramide |
| MALDI-TOF | matrix assisted laser desorption-ionization time-of-flight |
| MS | Mass Spectrometry |
| PBS | Phosphate Buffered Saline |
| PCR | Polymerase Chain Reaction |
| qPCR | Quantitative real-time PCR |
| SNA | Sambucus nigra agglutinin |
| WT | Wild-Type |
Conflicts of Interest
References
- Julien, S.; Bobowski, M.; Steenackers, A.; Le Bourhis, X.; Delannoy, P. How Do Gangliosides Regulate RTKs Signaling? Cells 2013, 2, 751–767. [Google Scholar] [CrossRef] [PubMed]
- Svennerholm, L. Ganglioside designation. Adv. Exp. Med. Biol. 1980, 125, 11–19. [Google Scholar] [PubMed]
- Yamashita, T.; Wada, R.; Sasaki, T.; Deng, C.; Bierfreund, U.; Sandhoff, K.; Proia, R.L. A vital role for glycosphingolipid synthesis during development and differentiation. Proc. Natl. Acad. Sci. USA 1999, 96, 9142–9147. [Google Scholar] [CrossRef]
- Ariga, T.; McDonald, M.P.; Yu, R.K. Role of ganglioside metabolism in the pathogenesis of Alzheimer’s disease—A review. J. Lipid Res. 2008, 49, 1157–1175. [Google Scholar] [CrossRef] [PubMed]
- Shahrizaila, N.; Yuki, N. Guillain-Barré syndrome animal model: The first proof of molecular mimicry in human autoimmune disorder. J. Biomed. Biotechnol. 2011, 2011. [Google Scholar] [CrossRef] [PubMed]
- Bobowski, M.; Cazet, A.; Steenackers, A.; Delannoy, P. Role of Complex Gangliosides in Cancer Progression. Carbohydr. Chem. 2012, 37, 1–20. [Google Scholar]
- Taki, T.; Hirabayashi, Y.; Ishikawa, H.; Ando, S.; Kon, K.; Tanaka, Y.; Matsumoto, M. A ganglioside of rat ascites hepatoma AH 7974F cells. Occurrence of a novel disialoganglioside (GD1 alpha) with a unique N-acetylneuraminosyl (alpha 2–6)-N-acetylgalactosamine structure. J. Biol. Chem. 1986, 261, 3075–3078. [Google Scholar] [PubMed]
- Hirabayashi, Y.; Hyogo, A.; Nakao, T.; Tsuchiya, K.; Suzuki, Y.; Matsumoto, M.; Kon, K.; Ando, S. Isolation and characterization of extremely minor gangliosides; GM1b and GD1 alpha; in adult bovine brains as developmentally regulated antigens. J. Biol. Chem. 1990, 265, 8144–8151. [Google Scholar] [PubMed]
- Furuya, S.; Irie, F.; Hashikawa, T.; Nakazawa, K.; Kozakai, A.; Hasegawa, A.; Sudo, K.; Hirabayashi, Y. Ganglioside GD1 alpha in cerebellar Purkinje cells. Its specific absence in mouse mutants with Purkinje cell abnormality and altered immunoreactivity in response to conjunctive stimuli causing long-term desensitization. J. Biol. Chem. 1994, 269, 32418–32425. [Google Scholar] [PubMed]
- Harduin-Lepers, A. Vertebrate Sialyltransferases. In Sialobiology: Structure, Biosynthesis and Function. Sialic Acid Glycoconjugates in Health and Disease; Martínez-Duncker, J.T., Ed.; Bentham Science Publishers: Sharjah, United Arab Emirates, 2013; pp. 139–187. [Google Scholar]
- Okajima, T.; Fukumoto, S.; Ito, H.; Kiso, M.; Hirabayashi, Y.; Urano, T.; Furukawa, K. Molecular cloning of brain-specific GD1alpha synthase (ST6GalNAc V) containing CAG/Glutamine repeats. J. Biol. Chem. 1999, 274, 30557–30562. [Google Scholar] [CrossRef] [PubMed]
- Ikehara, Y.; Shimizu, N.; Kono, M.; Nishihara, S.; Nakanishi, H.; Kitamura, T.; Narimatsu, H.; Tsuji, S.; Tatematsu, M. A novel glycosyltransferase with a polyglutamine repeat; a new candidate for GD1alpha synthase (ST6GalNAc V). FEBS Lett. 1999, 463, 92–96. [Google Scholar] [CrossRef] [PubMed]
- Okajima, T.; Chen, H.H.; Ito, H.; Kiso, M.; Tai, T.; Furukawa, K.; Urano, T.; Furukawa, K. Molecular cloning and expression of mouse GD1alpha/GT1aalpha/GQ1balpha synthase (ST6GalNAc VI) gene. J. Biol. Chem. 2000, 275, 6717–6723. [Google Scholar] [CrossRef] [PubMed]
- Tsuchida, A.; Okajima, T.; Furukawa, K.; Ando, T.; Ishida, H.; Yoshida, A.; Nakamura, Y.; Kannagi, R.; Kiso, M.; Furukawa, K. Synthesis of disialyl Lewis a (Le(a)) structure in colon cancer cell lines by a sialyltransferase; ST6GalNAc VI; responsible for the synthesis of alpha-series gangliosides. J. Biol. Chem. 2003, 278, 22787–22794. [Google Scholar] [CrossRef] [PubMed]
- Kroes, R.A.; He, H.; Emmett, M.R.; Nilsson, C.L.; Leach, F.E., 3rd; Amster, I.J.; Marshall, A.G.; Moskal, J.R. Overexpression of ST6GalNAcV, a ganglioside-specific alpha2,6-sialyltransferase, inhibits glioma growth in vivo. Proc. Natl. Acad. Sci. USA 2010, 107, 12646–12651. [Google Scholar] [CrossRef] [PubMed]
- Taki, T.; Ishikawa, D.; Ogura, M.; Nakajima, M.; Handa, S. Ganglioside GD1alpha functions in the adhesion of metastatic tumor cells to endothelial cells of the target tissue. Cancer Res. 1997, 57, 1882–1888. [Google Scholar] [PubMed]
- Chu, C.; Lugovtsev, V.; Golding, H.; Betenbaugh, M.; Shiloach, J. Conversion of MDCK cell line to suspension culture by transfecting with human siat7e gene and its application for influenza virus production. Proc. Natl. Acad. Sci. USA 2009, 106, 14802–14807. [Google Scholar] [CrossRef] [PubMed]
- Jaluria, P.; Betenbaugh, M.; Konstantopoulos, K.; Frank, B.; Shiloach, J. Application of microarrays to identify and characterize genes involved in attachment dependence in HeLa cells. Metab. Eng. 2007, 9, 241–251. [Google Scholar] [CrossRef] [PubMed]
- Bos, P.D.; Zhang, X.H.; Nadal, C.; Shu, W.; Gomis, R.R.; Nguyen, D.X.; Minn, A.J.; van de Vijver, M.J.; Gerald, W.L.; Foekens, J.A.; et al. Genes that mediate breast cancer metastasis to the brain. Nature 2009, 459, 1005–1009. [Google Scholar] [CrossRef] [PubMed]
- Lorusso, G.; Rüegg, C. New insights into the mechanisms of organ-specific breast cancer metastasis. Semin. Cancer Biol. 2012, 22, 226–233. [Google Scholar] [CrossRef] [PubMed]
- Pfaffl, M.W. A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 2001, 29, e45. [Google Scholar] [CrossRef] [PubMed]
- Shibuya, N.; Goldstein, I.J.; Broekaert, W.F.; Nsimba-Lubaki, M.; Peeters, B.; Peumans, W.J. The elderberry (Sambucus nigra L.) bark lectin recognizes the Neu5Ac(alpha2–6)Gal/GalNAc sequence. J. Biol. Chem. 1987, 262, 1596–1601. [Google Scholar] [PubMed]
- Smith, D.F.; Song, X.; Cummings, R.D. Use of glycan microarrays to explore specificity of glycan-binding proteins. Methods Enzymol. 2010, 480, 417–444. [Google Scholar] [PubMed]
- Cazet, A.; Bobowski, M.; Rombouts, Y.; Lefebvre, J.; Steenackers, A.; Popa, I.; Guérardel, Y.; Le Bourhis, X.; Tulasne, D.; Delannoy, P. The ganglioside G(D2) induces the constitutive activation of c-Met in MDA-MB-231 breast cancer cells expressing the G(D3) synthase. Glycobiology 2012, 22, 806–816. [Google Scholar] [CrossRef] [PubMed]
- Steenackers, A.; Cazet, A.; Bobowski, M.; Rombouts, Y.; Lefebvre, J.; Guérardel, Y.; Tulasne, D.; Le Bourhis, X.; Delannoy, P. Expression of GD3 synthase modifies ganglioside profile and increases migration of MCF-7 breast cancer cells. Comptes Rendus Chim. 2012, 15, 3–14. [Google Scholar] [CrossRef]
- Domon, B.; Costello, C.E. Structure elucidation of glycosphingolipids and gangliosides using high-performance tandem mass spectrometry. Biochemistry 1988, 27, 1534–1543. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Ding, L.; Sandford, A.J. Selection of reference genes for gene expression studies in human neutrophils by real-time PCR. BMC Mol. Biol. 2005, 6. [Google Scholar] [CrossRef]
- Schnaar, R.L. Isolation of glycosphingolipids. Methods Enzymol 1994, 230, 348–370. [Google Scholar] [PubMed]
- Ciucanu, I.; Kerek, F. Rapid and simultaneous methylation of fatty and hydroxy fatty acids for gas-liquid chromatographic analysis. J. Chromatogr. A 1984, 284, 179–185. [Google Scholar] [CrossRef]
- Krengel, U.; Bousquet, P.A. Molecular recognition of gangliosides and their potential for cancer immunotherapies. Front. Immunol. 2014, 5. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are not available.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Vandermeersch, S.; Vanbeselaere, J.; Delannoy, C.P.; Drolez, A.; Mysiorek, C.; Guérardel, Y.; Delannoy, P.; Julien, S. Accumulation of GD1α Ganglioside in MDA-MB-231 Breast Cancer Cells Expressing ST6GalNAc V. Molecules 2015, 20, 6913-6924. https://doi.org/10.3390/molecules20046913
Vandermeersch S, Vanbeselaere J, Delannoy CP, Drolez A, Mysiorek C, Guérardel Y, Delannoy P, Julien S. Accumulation of GD1α Ganglioside in MDA-MB-231 Breast Cancer Cells Expressing ST6GalNAc V. Molecules. 2015; 20(4):6913-6924. https://doi.org/10.3390/molecules20046913
Chicago/Turabian StyleVandermeersch, Sandy, Jorick Vanbeselaere, Clément P. Delannoy, Aurore Drolez, Caroline Mysiorek, Yann Guérardel, Philippe Delannoy, and Sylvain Julien. 2015. "Accumulation of GD1α Ganglioside in MDA-MB-231 Breast Cancer Cells Expressing ST6GalNAc V" Molecules 20, no. 4: 6913-6924. https://doi.org/10.3390/molecules20046913
APA StyleVandermeersch, S., Vanbeselaere, J., Delannoy, C. P., Drolez, A., Mysiorek, C., Guérardel, Y., Delannoy, P., & Julien, S. (2015). Accumulation of GD1α Ganglioside in MDA-MB-231 Breast Cancer Cells Expressing ST6GalNAc V. Molecules, 20(4), 6913-6924. https://doi.org/10.3390/molecules20046913