Development of a Novel Antimicrobial Screening System Targeting the Pyoverdine-Mediated Iron Acquisition System and Xenobiotic Efflux Pumps
Abstract
:1. Introduction
2. Results
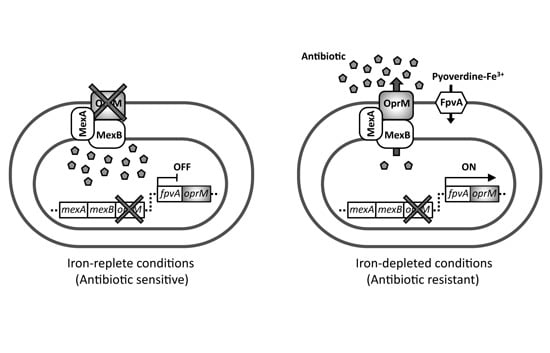
2.1. Experimental Design for Screening of Inhibitors toward Pyoverdine-Mediated Iron-Acquisition System and Xenobiotic Efflux Pumps

2.2. Antibiotic Susceptibility of the Test Strain Depends on the Extracellular Iron Levels
| Strain | Plasmid | Mic (µg/mL) | |||
|---|---|---|---|---|---|
| Aztreonam | Chloramphenicol | ||||
| Fe-replete | Fe-depleted | Fe-replete | Fe-depleted | ||
| PAO4290 (parent) | - | 4.0 | 4.0 | 32.0 | 32.0 |
| TNP072 (∆oprM) | - | 0.25 | 0.25 | 2.0 | 2.0 |
| TNP072 (∆oprM) | pOPRM1 | 4.0 | 4.0 | 32.0 | 32.0 |
| TNP091 (fpvA::oprM) | - | 0.25 | 2.0 | 4.0 | 16.0 |
| TNP092 (fpvA::oprM ∆pvdS) | - | 0.25 | 0.5 | 4.0 | 4.0 |
| TNP092 (fpvA::oprM ∆pvdS) | pPvdS | 4.0 | 4.0 | 32.0 | 32.0 |

2.3. The pvdS Mutation Prevents Induction of the oprM Expression

2.4. Screening of Inhibitors of the Iron-Acquisition System or the Efflux Pump in a Chemical Library

2.5. Characterization of the Hit-Compound


3. Discussion

4. Experimental Section
4.1. Bacterial Strains, Plasmids, and Growth Conditions
| Strains or Plasmids | Relevant Property | Source or Reference |
|---|---|---|
| Strain | ||
| Escherichia coli JM109 | recA1, endA1, gyrA96, thi, hsdR17(rk− mk+), e14−(mcrA−), supE44, relA1, Δ(lac-proAB)/ F'[traD36, proAB+, lacIq, lacZΔM15] | Laboratory strain |
| Escherichia coli S17-1 | RP4-2-Tc::Mu-Km::Tn7, pro, res−, mod+ | [28] |
| Escherichia coli MG1655ΔBC | ΔacrB, ΔtolC | [29] |
| Escherichia coli BW25113 | Wild type, parent of JW0451 | [30] |
| Escherichia coli JW0451 | ΔacrB derived from BW25113 | [30] |
| Pseudomonas aeruginosa PAO1 | wild type | Laboratory strain |
| Pseudomonas aeruginosa PAO4290 | leu-10, argF10, aph-9004, FP- | Matsumoto collection |
| Pseudomonas aeruginosa TNP072 | PAO4290 ΔoprM::Tcr | [20] |
| Pseudomonas aeruginosa TNP091 | TNP072 fpvA::oprM | This study |
| Pseudomonas aeruginosa TNP092 | TNP072 fpvA::oprM, ΔpvdS | This study |
| Pseudomonas aeruginosa TNP30#1 | PAO4290-derived multiantibiotic resistant mutant overexpressing mexAB-OprM genes | [31] |
| Plasmid | ||
| pK19mobsacB | pK19 derivative carrying RP4 mob region and Bacillus subtilis sacB gene, Kmr | [32] |
| pK18mob | pK18 derivative carrying RP4 mob region, Kmr | [32] |
| pMMB67EH | Broad host range vector, Apr, IncQ | [24] |
| pMMB67HE | Broad host range vector, Apr, IncQ | [24] |
| pK19FpvOprM | pK19mobsacB derivative carrying 3'-end of fpvA region and its downstream region in which oprM gene is inserted | This study |
| pK18mobPvdS | pK18mob derivative caring internal region of the pvdS gene | This study |
| pOPRM1 | pMMB67EH derivative carrying wild type oprM gene, Apr | [21] |
| pPvdS | pMMB67EH derivative carrying the pvdS gene | This study |
| pABM | pMMB67HE derivative carrying the mexA-mexB-oprM genes | [29] |
4.2. Construction of the Recombinant Strain
| Primer | Nucleotide sequence a | Tag |
|---|---|---|
| FpvAdown-F | 5′-GGCCCCGGGGCCCGACTGCGAAAAAC-3′ | SmaI |
| FpvAdown-R | 5′-CGAATTCGCACGCAACTGGTGGGATAC-3′ | EcoRI |
| OprM-F | 5′-TGTCTAGACAAGCAGCAGGCGTCCGTC-3′ | XbaI |
| OprM-R | 5′-TGACCCGGGTCAAGCCTGGGGATCTTCC-3' | SmaI |
| FpvA-F | 5′-ATTAAGCTTAATCCCGACACCATGCTTAC-3′ | HindIII |
| FpvA-R | 5′-GATCTAGATCAGAAGTCCCAGCGAGTG-3′ | XbaI |
| PvdS-F | 5′-TGAATTCACCGTACGATCCTGGTGAAG-3′ | EcoRI |
| PvdS-R | 5′-TGTCTAGACATGAAGTTGACCAGGGTCG-3′ | XbaI |
4.3. Expression of OprM under Iron-Replete and Iron-Depleted Conditions
4.4. Screening of a Chemical Library
4.5. Assay for the Potentiation of Antibiotic Activity by a Hit-Compound
4.6. Other Techniques
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Boucher, H.W.; Talbot, G.H.; Bradley, J.S.; Edwards, J.E.; Gilbert, D.; Rice, L.B.; Scheld, M.; Spellberg, B.; Bartlett, J. Bad bugs, no drugs: No ESKAPE! An update from the Infectious Diseases Society of America. Clin. Infect. Dis. 2009, 48, 1–12. [Google Scholar] [CrossRef]
- Taubes, G. The bacteria fight back. Science 2008, 321, 356–361. [Google Scholar] [CrossRef] [PubMed]
- Hunter, P. Where next for antibiotics? The immune system and the nature of pathogenicity are providing vital clues in the fight against antibiotic-resistant bacteria. EMBO Rep. 2012, 13, 680–683. [Google Scholar]
- Bush, K.; Courvalin, P.; Dantas, G.; Davies, J.; Eisenstein, B.; Huovinen, P.; Jacoby, G.A.; Kishony, R.; Kreiswirth, B.N.; Kutter, E.; et al. Tackling antibiotic resistance. Nat. Rev. Microbiol. 2011, 9, 894–896. [Google Scholar] [CrossRef]
- Fischbach, M.A.; Walsh, C.T. Antibiotics for emerging pathogens. Science 2009, 325, 1089–1093. [Google Scholar] [CrossRef] [PubMed]
- Payne, D.J.; Gwynn, M.N.; Holmes, D.J.; Pompliano, D.L. Drugs for bad bugs: Confronting the challenges of antibacterial discovery. Nat. Rev. Drug Discov. 2007, 6, 29–40. [Google Scholar] [CrossRef] [PubMed]
- Taylor, P.L.; Wright, G.D. Novel approaches to discovery of antibacterial agents. Anim. Health Res. Rev. 2008, 9, 237–246. [Google Scholar] [CrossRef] [PubMed]
- Gwynn, M.N.; Portnoy, A.; Rittenhouse, S.F.; Payne, D.J. Challenges of antibacterial discovery revisited. Ann. N. Y. Acad. Sci. 2010, 1213, 5–19. [Google Scholar] [CrossRef] [PubMed]
- Rasko, D.A.; Sperandio, V. Anti-virulence strategies to combat bacteria-mediated disease. Nat. Rev. Drug Discov. 2010, 9, 117–128. [Google Scholar] [CrossRef] [PubMed]
- Baron, C. Antivirulence drugs to target bacterial secretion systems. Curr. Opin. Microbiol. 2010, 13, 100–105. [Google Scholar] [CrossRef] [PubMed]
- Alekshun, M.N.; Levy, S.B. Targeting virulence to prevent infection: To kill or not to kill? Drug Discov. Today Ther. Strategies 2004, 1, 483–489. [Google Scholar] [CrossRef]
- Andrews, S.C.; Robinson, A.K.; Rodriguez-Quinones, F. Bacterial iron homeostasis. FEMS Microbiol. Rev. 2003, 27, 215–237. [Google Scholar] [CrossRef] [PubMed]
- Miethke, M.; Marahiel, M.A. Siderophore-based iron acquisition and pathogen control. Microbiol Mol. Biol. Rev. 2007, 71, 413–451. [Google Scholar] [CrossRef] [PubMed]
- Drakesmith, H.; Prentice, A.M. Hepcidin and the iron-infection axis. Science 2012, 338, 768–772. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, P. Iron uptake and metabolism in pseudomonads. Appl. Microbiol. Biotechnol. 2010, 86, 1637–1645. [Google Scholar] [CrossRef] [PubMed]
- Beare, P.A.; For, R.J.; Martin, L.W.; Lamont, I.L. Siderophore-mediated cell signalling in Pseudomonas aeruginosa: Divergent pathways regulate virulence factor production and siderophore receptor synthesis. Mol. Microbiol. 2003, 47, 195–207. [Google Scholar] [CrossRef] [PubMed]
- Lamont, I.L.; Beare, P.A.; Ochsner, U.; Vasil, A.I.; Vasil, M.L. Siderophore-mediated signaling regulates virulence factor production in Pseudomonas aeruginosa. Proc. Natl. Acad. Sci. USA 2002, 99, 7072–7077. [Google Scholar] [CrossRef] [PubMed]
- Cornelis, P.; Matthijs, S.; Van Oeffelen, L. Iron uptake regulation in Pseudomonas aeruginosa. Biometals 2009, 22, 15–22. [Google Scholar] [CrossRef] [PubMed]
- Voulhoux, R.; Filloux, A.; Schalk, I.J. Pyoverdine-mediated iron uptake in Pseudomonas aeruginosa: The Tat system is required for PvdN but not for FpvA transport. J. Bacteriol. 2006, 188, 3317–3323. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, H.; Ocaktan, A.; Tsuda, M.; Nakae, T. The role of mex-gene products in antibiotic extrusion in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 1997, 233, 611–618. [Google Scholar] [CrossRef] [PubMed]
- Yoneyama, H.; Ocaktan, A.; Gotoh, N.; Nishino, T.; Nakae, T. Subunit swapping in the Mex-extrusion pumps in Pseudomonas aeruginosa. Biochem. Biophys. Res. Commun. 1998, 244, 898–902. [Google Scholar] [CrossRef] [PubMed]
- Aires, J.R.; Kohler, T.; Nikaido, H.; Plesiat, P. Involvement of an active efflux system in the natural resistance of Pseudomonas aeruginosa to aminoglycosides. Antimicrob. Agents Chemother. 1999, 43, 2624–2628. [Google Scholar] [PubMed]
- Poole, K.; Neshat, S.; Heinrichs, D. Pyoverdine-mediated iron transport in Pseudomonas aeruginosa: Involvement of a high-molecular-mass outer membrane protein. FEMS Microbiol. Lett. 1991, 62, 1–5. [Google Scholar] [PubMed]
- Furste, J.P.; Pansegrau, W.; Frank, R.; Blocker, H.; Scholz, P.; Bagdasarian, M.; Lanka, E. Molecular cloning of the plasmid RP4 primase region in a multi-host-range tacP expression vector. Gene 1986, 48, 119–131. [Google Scholar] [CrossRef] [PubMed]
- Piddock, L.J.; Garvey, M.I.; Rahman, M.M.; Gibbons, S. Natural and synthetic compounds such as trimethoprim behave as inhibitors of efflux in Gram-negative bacteria. J. Antimicrob. Chemother. 2010, 65, 1215–1223. [Google Scholar] [CrossRef] [PubMed]
- Stavri, M.; Piddock, L.J.; Gibbons, S. Bacterial efflux pump inhibitors from natural sources. J. Antimicrob. Chemother. 2007, 59, 1247–1260. [Google Scholar] [CrossRef] [PubMed]
- Ochsner, U.A.; Snyder, A.; Vasil, A.I.; Vasil, M.L. Effects of the twin-arginine translocase on secretion of virulence factors, stress response, and pathogenesis. Proc. Natl. Acad. Sci. USA 2002, 99, 8312–8317. [Google Scholar] [CrossRef] [PubMed]
- Simon, R.; Priefer, U.; Puhler, A. A Broad Host Range Mobilization System for In Vivo Genetic Engineering: Transposon Mutagenesis in Gram Negative Bacteria. Nat. Biotech. 1983, 1, 784–791. [Google Scholar] [CrossRef]
- Matsumoto, Y.; Hayama, K.; Sakakihara, S.; Nishino, K.; Noji, H.; Iino, R.; Yamaguchi, A. Evaluation of multidrug efflux pump inhibitors by a new method using microfluidic channels. PLoS ONE 2011, 6, e18547. [Google Scholar] [CrossRef] [PubMed]
- Baba, T.; Ara, T.; Hasegawa, M.; Takai, Y.; Okumura, Y.; Baba, M.; Datsenko, K.A.; Tomita, M.; Wanner, B.L.; Mori, H. Construction of Escherichia coli K-12 in-frame, single-gene knockout mutants: The Keio collection. Mol. Syst. Biol. 2006, 2, 2006.0008. [Google Scholar] [CrossRef]
- Saito, K.; Yoneyama, H.; Nakae, T. nalB-type mutations causing the overexpression of the MexAB-OprM efflux pump are located in the mexR gene of the Pseudomonas aeruginosa chromosome. FEMS Microbiol. Lett. 1999, 179, 67–72. [Google Scholar] [CrossRef] [PubMed]
- Schafer, A.; Tauch, A.; Jager, W.; Kalinowski, J.; Thierbach, G.; Puhler, A. Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: Selection of defined deletions in the chromosome of Corynebacterium glutamicum. Gene 1994, 145, 69–73. [Google Scholar] [CrossRef] [PubMed]
- Akiba, K.; Ando, T.; Isogai, E.; Nakae, T.; Yoneyama, H. Tat pathway-mediated translocation of the Sec pathway substrate OprM, an outer membrane subunit of the resistance nodulation division xenobiotic extrusion pumps, in Pseudomonas aeruginosa. Chemotherapy 2013, 59, 129–137. [Google Scholar] [CrossRef] [PubMed]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Lowry, O.H.; Rosebrough, N.J.; Farr, A.L.; Randall, R.J. Protein measurement with the Folin phenol reagent. J. Biol. Chem. 1951, 193, 265–275. [Google Scholar] [PubMed]
- Sanger, F.; Nicklen, S.; Coulson, A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. USA 1977, 74, 5463–5467. [Google Scholar] [CrossRef] [PubMed]
- Sample Availability: Not available.
© 2015 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, K.; Ushioda, K.; Akiba, K.; Matsumoto, Y.; Maseda, H.; Ando, T.; Isogai, E.; Nakae, T.; Yoneyama, H. Development of a Novel Antimicrobial Screening System Targeting the Pyoverdine-Mediated Iron Acquisition System and Xenobiotic Efflux Pumps. Molecules 2015, 20, 7790-7806. https://doi.org/10.3390/molecules20057790
Sato K, Ushioda K, Akiba K, Matsumoto Y, Maseda H, Ando T, Isogai E, Nakae T, Yoneyama H. Development of a Novel Antimicrobial Screening System Targeting the Pyoverdine-Mediated Iron Acquisition System and Xenobiotic Efflux Pumps. Molecules. 2015; 20(5):7790-7806. https://doi.org/10.3390/molecules20057790
Chicago/Turabian StyleSato, Kazuki, Kenichi Ushioda, Keiji Akiba, Yoshimi Matsumoto, Hideaki Maseda, Tasuke Ando, Emiko Isogai, Taiji Nakae, and Hiroshi Yoneyama. 2015. "Development of a Novel Antimicrobial Screening System Targeting the Pyoverdine-Mediated Iron Acquisition System and Xenobiotic Efflux Pumps" Molecules 20, no. 5: 7790-7806. https://doi.org/10.3390/molecules20057790
APA StyleSato, K., Ushioda, K., Akiba, K., Matsumoto, Y., Maseda, H., Ando, T., Isogai, E., Nakae, T., & Yoneyama, H. (2015). Development of a Novel Antimicrobial Screening System Targeting the Pyoverdine-Mediated Iron Acquisition System and Xenobiotic Efflux Pumps. Molecules, 20(5), 7790-7806. https://doi.org/10.3390/molecules20057790