Antimicrobial Activity and Urease Inhibition of Schiff Bases Derived from Isoniazid and Fluorinated Benzaldehydes and of Their Copper(II) Complexes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis
2.2. Analytical Characterization of the Compounds
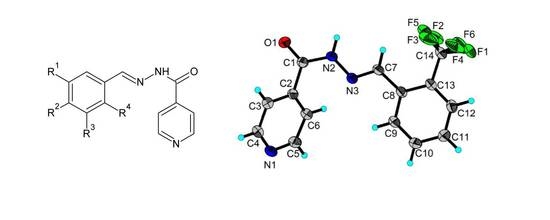
2.3. X-ray Structures of the Schiff Bases
2.4. Bioactivity of the Ligands and Complexes
3. Materials and Methods
3.1. General
3.2. Synthesis of the Schiff Bases
3.3. Synthesis of the Complexes
3.4. X-ray Single-Crystal Structure Determination
3.5. Urease Inhibition Assay
3.6. Determination of Antimicrobial Activity
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jakupec, M.A.; Galanski, M.; Arion, V.B.; Hartinger, C.; Keppler, B.K. Antitumour metal compounds: More than theme and variations. Dalton Trans. 2008, 183, 183–194. [Google Scholar] [CrossRef] [PubMed]
- Louie, A.Y.; Meade, T.J. Metal complexes as enzyme inhibitors. Chem. Rev. 1999, 99, 2711–2734. [Google Scholar] [CrossRef] [PubMed]
- Che, C.M.; Siu, F.M. Metal complexes in medicine with a focus on enzyme inhibition. Curr. Opin. Chem. Biol. 2010, 14, 255–261. [Google Scholar] [CrossRef] [PubMed]
- Kilpin, K.J.; Dyson, P.J. Enzyme inhibition by metal complexes: Concepts, strategies and applications. Chem. Sci. 2013, 4, 1410–1419. [Google Scholar] [CrossRef]
- Qin, Y.; Cabral, J.M.S. Properties and applications of urease. Biocatal. Biotransform. 2002, 20, 1–14. [Google Scholar] [CrossRef]
- Burne, R.A.; Chen, Y.Y. Bacterial ureases in infectious diseases. Microbes Infect. 2000, 2, 533–542. [Google Scholar] [CrossRef]
- Konieczna, I.; Zarnowiec, P.; Kwinkowski, M.; Kolesinska, B.; Fraczyk, J.; Kaminski, Z.; Kaca, W. Bacterial urease and its role in long-lasting human diseases. Curr. Protein Pept. Sci. 2012, 13, 789–806. [Google Scholar] [CrossRef] [PubMed]
- Rutherford, J.C. The emerging role of urease as a general microbial virulence factor. PLoS Pathog. 2014, 10, e1004062. [Google Scholar] [CrossRef] [PubMed]
- Mobley, H.L. The role of Helicobacter pylori urease in the pathogenesis of gastritis and peptic ulceration. Aliment. Pharmacol. Ther. 1996, 10 (Suppl. 1), 57–64. [Google Scholar] [CrossRef] [PubMed]
- Ashwini, P.; Sumana, M.N.; Shilpa, U.; Mamatha, P.; Manasa, P.; Dhananjaya, B.L.; Farhan, Z.; Nagendra, P.M.N. A review on Helicobacter pylori: Its biology, complications and management. Int. J. Pharm. Pharm. Sci. 2015, 7 (Suppl. 1), 14–20. [Google Scholar]
- Marshall, B. Helicobacter connections. ChemMedChem 2006, 1, 783–802. [Google Scholar] [CrossRef] [PubMed]
- Gouzy, A.; Poquet, Y.; Neyrolles, O. Nitrogen metabolism in Mycobacterium tuberculosis physiology and virulence. Nat. Rev. Microbiol. 2014, 12, 729–737. [Google Scholar] [CrossRef] [PubMed]
- Ibrar, A.; Khan, I.; Abbas, N. Structurally diversified heterocycles and related privileged scaffolds as potential urease inhibitors: A brief overview. Arch. Pharm. Chem. Life Sci. 2013, 346, 423–446. [Google Scholar] [CrossRef] [PubMed]
- Zaborska, W.; Krajewska, B.; Olech, Z. Heavy metal ions inhibition of Jack bean urease: Potential for rapid contaminant probing. J. Enzyme Inhib. Med. Chem. 2004, 19, 65–69. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, B. Ureases I. Functional, catalytic and kinetic properties: A review. J. Mol. Catal. B Enzym. 2009, 59, 9–21. [Google Scholar] [CrossRef]
- Habala, L.; Roller, A.; Matuška, M.; Valentová, J.; Rompel, A.; Devínsky, F. Complexes of N-hydroxyethyl-N-benzimidazolylmethylethylenediaminediacetic acid with copper(II) and cobalt(II): Preparation, crystal structure and urease inhibitory activity. Inorg. Chim. Acta 2014, 421, 423–426. [Google Scholar] [CrossRef]
- Křikavová, R.; Vančo, J.; Trávníček, Z.; Buchtík, R.; Dvořák, Z. Copper(II) quinolinato-7-carboxamido complexes as potent antitumor agents with broad spectra and selective effects. RSC Adv. 2016, 6, 3899–3909. [Google Scholar] [CrossRef]
- Habala, L.; Bartel, C.; Giester, G.; Ionita, G.; Jakupec, M.; Valentová, J.; Keppler, B.K.; Rompel, A.; Devínsky, F. Preparation, characterization and bioactivity of a novel oxovanadium complex. New trends in coordination, bioinorganic, and applied inorganic chemistry. In Monograph Series of the International Conferences on Coordination Chemistry held Periodically at Smolenice in Slovakia; Slovak University of Technology Press: Bratislava, Slovakia, 2011; Volume 11, pp. 124–132. [Google Scholar]
- Habala, L.; Bartel, C.; Giester, G.; Jakupec, M.A.; Keppler, B.K.; Rompel, A. Complexes of N-hydroxyethyl-N-benzimidazolylmethylethylenediaminediacetic acid with group 12 metals and vanadium—Synthesis, structure and bioactivity of the vanadium complex. J. Inorg. Biochem. 2015, 147, 147–152. [Google Scholar] [CrossRef] [PubMed]
- Böhm, H.J.; Banner, D.; Bendels, S.; Kansy, M.; Kuhn, B.; Müller, K.; Obst-Sander, U.; Stahl, M. Fluorine in medicinal chemistry. ChemBioChem 2004, 5, 637–643. [Google Scholar] [CrossRef] [PubMed]
- Qin, W.; Long, S.; Panunzio, M.; Biondi, S. Schiff bases: A short survey on an evergreen chemistry tool. Molecules 2013, 18, 12264–12289. [Google Scholar] [CrossRef] [PubMed]
- Utreja, D.V.; Singh, S.; Kaur, M. Schiff bases and their metal complexes as anti-cancer agents: A review. Curr. Bioact. Compd. 2015, 11, 215–230. [Google Scholar] [CrossRef]
- Bottari, B.; Maccari, R.; Monforte, F.; Ottanà, R.; Rotondo, E.; Vigorita, M.G. Isoniazid-related copper(II) and nickel(II) complexes with antimycobacterial in vitro activity. Bioorg. Med. Chem. Lett. 2000, 10, 657–660. [Google Scholar] [CrossRef]
- Bottari, B.; Maccari, R.; Monforte, F.; Ottanà, R.; Rotondo, E.; Vigorita, M.G. Antimycobacterial in vitro activity of cobalt(II) isonicotinoylhydrazone complexes. Bioorg. Med. Chem. Lett. 2001, 11, 301–303. [Google Scholar] [CrossRef]
- Maccari, R.; Ottanà, R.; Bottari, B.; Rotondo, E.; Vigorita, M.G. In vitro advanced antimycobacterial screening of cobalt(II) and copper(II) complexes of fluorinated isonicotinoylhydrazones. Bioorg. Med. Chem. Lett. 2004, 14, 5731–5733. [Google Scholar] [CrossRef] [PubMed]
- Sharma, K.K.; Singh, R.; Fahmi, N.; Singh, R.V. Synthesis, coordination behavior, and investigations of pharmacological effects of some transition metal complexes with isoniazid Schiff bases. J. Coord. Chem. 2010, 63, 3071–3082. [Google Scholar] [CrossRef]
- Poggi, M.; Barroso, R.; Costa-Filho, A.J.; de Barros, H.B.; Pavan, F.; Leite, C.Q.; Gambino, D.; Torre, M.H. New isoniazid complexes, promising agents against Mycobacterium tuberculosis. J. Mex. Chem. Soc. 2013, 57, 198–204. [Google Scholar]
- Judge, V.; Narasimhan, B.; Ahuja, M. Isoniazid: the magic molecule. Med. Chem. Res. 2012, 21, 3940–3957. [Google Scholar] [CrossRef]
- Ababei, L.V.; Kriza, A.; Andronescu, C.; Musuc, A.M. Synthesis and characterization of new complexes of some divalent transition metals with N-isonicotinamido-4-chlorobenzalaldimine. J. Serb. Chem. Soc. 2011, 76, 1103–1115. [Google Scholar] [CrossRef]
- Tayamon, S.; Mazlan, N.A.; Ravoof, T.B.S.A.; Tahir, M.I.M.; Crouse, K.A. 2-Cyclohexylidene-N-methylhydrazinecarbothioamide. Acta Cryst. E 2012, 68, o3104–o3105. [Google Scholar] [CrossRef] [PubMed]
- Tecer, E.; Dege, N.; Zülfikaroğlu, A.; Şenyüzb, N.; Batıb, H. N′-(2,3-Dihydroxybenzylidene)-isonicotinohydrazide. Acta Cryst. E 2010, 66, o3369–o3370. [Google Scholar] [CrossRef] [PubMed]
- Zülfikaroğlu, A.; Yüksektepe, Ç.; Bati, H.; Çalışkan, N.; Büyükgüngör, O. Crystal structure and properties of (Z)-N′-((E)-2-(hydroxyimino)-1-phenylethylidene) isonicotinohydrazide. J. Struct. Chem. 2009, 50, 1166–1170. [Google Scholar] [CrossRef]
- Li, H.B. 2′-(2-Hydroxy-3,5-diiodobenzylidene)-isonicotinohydrazide methanol solvate. Acta Cryst. E 2007, 63, o972–o973. [Google Scholar] [CrossRef]
- He-Bing, L. 3-Bromo-N′-(2-hydroxybenzylidene)-benzohydrazide. Acta Cryst. E 2008, 64, o465. [Google Scholar] [CrossRef]
- Puvaneswary, S.; Ali, H.M.; Robinson, W.T.; Ng, S.W. N′-(2-Hydroxy-5-nitrobenzylidene)-2-(1H-indol-3-yl)acetohydrazide. Acta Cryst. E 2008, E64, o1777. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.H.; Diao, Y.P.; Li, M.L.; Yan, X.; Liu, J. (E)-4-Bromo-N′-(2-chlorobenzylidene)-benzohydrazide. Acta Cryst. E 2009, 65, o1034. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Q.H.; Xu, Y.H.; Jian, L.Y.; Zhao, L.M. 3,5-Dihydroxy-N′-(2-hydroxybenzylidene) benzohydrazide monohydrate. Acta Cryst. E 2008, 64, o338. [Google Scholar] [CrossRef] [PubMed]
- Warad, I.; Haddad, S.F.; Al-Noaimi, M.; Hammouti, B.; Hadda, T.B. N′-[(E)-2-Chlorobenzylidene]thiophene-2-carbohydrazide. Acta Cryst. E 2013, 69, o1442. [Google Scholar] [CrossRef] [PubMed]
- Kargar, H.; Kia, R.; Tahir, M.N. (E)-4-Amino-N′-(5-bromo-2-hydroxybenzylidene)benzohydrazide monohydrate. Acta Cryst. E 2012, 68, o2120. [Google Scholar] [CrossRef] [PubMed]
- Ren, C.Y. (E)-6-Chloro-N′-(3,5-dichloro-2-hydroxybenzylidene)nicotinohydrazide. Acta Cryst. E 2009, 65, o678. [Google Scholar] [CrossRef] [PubMed]
- Kargar, H.; Kia, R.; Tahir, M.N. (E)-4-Amino-N′-(5-chloro-2-hydroxybenzylidene)benzohydrazide. Acta Cryst. E 2012, 68, o2118–o2119. [Google Scholar] [CrossRef] [PubMed]
- Khoo, T.J.; bin Break, M.K.; Crouse, K.A.; Tahir, M.I.M.; Ali, A.M.; Cowley, A.R.; Watkin, D.J.; Tarafder, M.T.H. Synthesis, characterization and biological activity of two Schiff base ligands and their nickel(II), copper(II), zinc(II) and cadmium(II) complexes derived from S-4-picolyldithiocarbazate and X-ray crystal structure of cadmium(II) complex derived from pyridine-2-carboxaldehyde. Inorg. Chim. Acta 2014, 413, 68–76. [Google Scholar]
- Ferguson, L.N.; Reid, J.C.; Calvin, M. Some new fluorinated phenolic aldehydes and acids. J. Am. Chem. Soc. 1946, 68, 2502–2504. [Google Scholar] [CrossRef]
- Oxford Diffraction. CrysAlis PRO and CrysAlisRed; Oxford Diffraction Ltd.: Abingdon, UK, 2016. [Google Scholar]
- Palatinus, L. The charge-flipping algorithm in crystallography. Acta Cryst. B 2013, 69, 1–16. [Google Scholar] [CrossRef]
- Dolomanov, O.V.; Bourhis, L.J.; Gildea, R.J.; Howard, J.A.K.; Puschmann, H. OLEX2: A complete structure solution, refinement and analysis program. J. Appl. Cryst. 2009, 42, 339–341. [Google Scholar] [CrossRef]
- Sheldrick, G.M. A short history of SHELX. Acta Cryst. A 2008, 64, 112–122. [Google Scholar] [CrossRef] [PubMed]
- Brandenburg, K.; Berndt, M. DIAMOND; Crystal Impact GbR: Bonn, Germany, 1999. [Google Scholar]
- Tanaka, T.; Kawase, M.; Tani, S. Urease inhibitory activity of simple α,β-unsaturated ketones. Life Sci. 2003, 73, 2985–2990. [Google Scholar] [CrossRef]
- Bilková, A.; Paulovičová, E.; Paulovičová, L.; Poláková, M. Antimicrobial activity of mannose-derived glycosides. Monatsh. Chem. 2015, 146, 1707–1714. [Google Scholar] [CrossRef]
- Sample Availability: Samples of the compounds are not available from the authors.






| Ligand | L1 | L2 | L3 | L4 |
|---|---|---|---|---|
| Bond distances | ||||
| C1–N2 | 1.3509(17) | 1.353(2) | 1.347(2) | 1.3500(17) |
| N2–N3 | 1.3800(15) | 1.3679(18) | 1.389(2) | 1.3786(14) |
| N3–C7 | 1.2690(18) | 1.270(2) | 1.273(3) | 1.2764(17) |
| Torsion angles | ||||
| C1–N2–N3–C7 | 169.57(13) | −173.01(16) | −171.94(19) | −176.96(11) |
| Ligand | D–H...A | d(D–H) | d(H...A) | d(D...A) | <(DHA) |
|---|---|---|---|---|---|
| L1 | C7–H7A...O1 i | 0.93 | 2.45 | 3.2591(16) | 145.4 |
| N2–H2A... O1 i | 0.86 | 2.15 | 3.9797(14) | 161.0 | |
| L2 | C7–H7...F2 | 0.93 | 2.46 | 2.980(2) | 115.4 |
| C7–H7...F5 | 0.93 | 2.10 | 2.757(13) | 126.6 | |
| N2–H2N...N1 i | 0.89(2) | 2.17(2) | 3.0482(18) | 172.5(19) | |
| L3 | O2–H2A...O1 | 0.8396(19) | 2.274(16) | 2.954(2) | 138(2) |
| O2–H2A...N3 | 0.8396(19) | 2.565(14) | 3.271(2) | 142(2) | |
| C7–H7...O2 i | 0.93 | 2.64 | 3.338(2) | 132.3 | |
| N2–H2...O2 i | 0.86 | 2.02 | 2.834(2) | 157.8 | |
| O2–H2B...N1 ii | 0.835(18) | 2.06(2) | 2.861(2) | 161(2) | |
| C4–H4...F1 iii | 0.93 | 2.55 | 3.349(3) | 144.6 | |
| L4 | O2–H2O...N3 | 0.82 | 1.92 | 2.6316(15) | 145.3 |
| C4–H4...F1 i | 0.93 | 2.64 | 3.281(2) | 126.9 | |
| C12–H12...O1 ii | 0.93 | 2.51 | 3.2566(18) | 138.1 | |
| N2–H2N...N1 iii | 0.86 | 2.19 | 2.8861(15) | 138.1 |
| Compound | S. aureus Mau 82/78 | E. coli 327/73 | C. Albicans 59/91 | |
|---|---|---|---|---|
| MIC [mM] | L1 | 37.5 | 9.37 | 0.037 |
| L2 | 37.5 | 18.76 | 9.37 | |
| L3 | n.i. | 25.0 | 25.0 | |
| L4 | n.i. | 1.55 | 0.048 | |
| L5 | 50.0 | 25.0 | 12.5 | |
| L6 | 50.0 | 12.5 | 12.5 | |
| Cu-L3 | 6.25 | 6.25 | 6.25 | |
| Cu-L1 | 6.25 | 12.5 | 6.25 | |
| Cu-L2 | 12.5 | 12.5 | 12.5 | |
| Cu-L6 | 0.76 | 6.25 | 6.25 | |
| Isoniazid | n.i. | n.i. | 50.0 | |
| Ciprofloxacin | 6.8 × 10−4 | <3 × 10−4 | n.a. |
| Compounds | IC50 ± SD (μM) |
|---|---|
| ligands | >500 |
| Cu-L3 | 1.17 ± 0.02 |
| Cu-L1 | 0.49 ± 0.01 |
| Cu-L2 | 1.25 ± 0.03 |
| Cu-L6 | 1.01 ± 0.02 |
| Acetohydroxamic acid | 185 ± 6.2 |
| L1 | L2 | L3 | L4 | |
|---|---|---|---|---|
| Empirical formula | C14H10F3N3O | C14H10F3N3O | C13H12FN3O2 | C13H10FN3O2 |
| Temperature (K) | 293(2) K | 293(2) K | 293(2) K | 293(2) K |
| Wavelength (Å) | 0.71073 | 1.54184 | 0.71073 | 0.71073 |
| Crystal system | Monoclinic | Monoclinic | Orthorhombic | Monoclinic |
| Space group | P21/n | P21/c | P212121 | P21/c |
| Unit cell dimensions | a = 11.7577(7) Å | a = 8.1335(2) Å | a = 6.4785(4) Å | a = 8.9048(3) Å |
| b = 5.1630(2) Å | b = 7.2718(2) Å | b = 7.0244(10) Å | b = 10.1067(3) Å | |
| c = 21.6108(13) Å | c = 22.2575(6) Å | c = 27.373(4) Å | c = 13.6087(5) Å | |
| a = 90° | a = 90° | a = 90° | a = 90° | |
| b = 98.143(5)° | b = 98.686(2)° | b = 90° | b = 103.420(4)° | |
| g = 90° | g = 90° | g = 90° | g = 90° | |
| Formula weight | 293.25 | 293.25 | 261.26 | 259.24 |
| Volume (Å3) | 1298.66(12) | 1301.33(6) | 1245.7(3) | 1191.32(7) |
| Z/Calculated density (mg/m3) | 4/1.500 | 4/1.497 | 4/1.393 | 4/1.445 |
| Absorption coefficient (mm−1) | 0.127 | 1.101 | 0.107 | 0.111 |
| F (000) | 600 | 600 | 544 | 536 |
| Crystal size (mm) | 0.97 × 0.78 × 0.21 | 0.30 × 0.15 × 0.05 | 0.37 × 0.11 × 0.08 | 0.62 × 0.23 × 0.12 |
| Theta range for data collection | 3.132° to 26.361° | 4.018° to 75.559° | 2.977° to 30.497° | 3.097° to 26.368° |
| Index ranges | −14 ≤ h ≤ 14 | −10 ≤ h ≤ 10 | −9 ≤ h ≤ 9 | −11 ≤ h ≤ 11 |
| −6 ≤ k ≤ 6 | −9 ≤ k ≤ 9 | −10 ≤ k ≤ 10 | −12 ≤ k ≤ 12 | |
| −27 ≤ l ≤ 27 | −27 ≤ l ≤ 27 | −39 ≤ l ≤ 39 | −16 ≤ l ≤ 16 | |
| Reflections collected | 19468 | 19144 | 27502 | 18517 |
| Independent reflections | 2654 [R(int) = 0.0218] | 2675 [R(int) = 0.0418] | 3797 [R(int) = 0.0352] | 2432 [R(int) = 0.0292] |
| Completeness to 2Θ = 25.000° | 99.7% | 100.0% | 99.9% | 99.8% |
| Refinement method | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 | Full-matrix least-squares on F2 |
| Data/restraints/parameters | 2654/268/218 | 2675/268/221 | 3797/3/178 | 2432/0/174 |
| Goodness-of-fit on F2 | 1.089 | 1.034 | 1.015 | 1.066 |
| Final R indices (I > 2σ(I)) | R1 = 0.0405 | R1 = 0.0410 | R1 = 0.0428 | R1 = 0.0382 |
| wR2 = 0.1063 | wR2 = 0.1133 | wR2 = 0.0946 | wR2 = 0.1035 | |
| R indices (all data) | R1 = 0.0474 | R1 = 0.0530 | R1 = 0.0633 | R1 = 0.0461 |
| wR2 = 0.1120 | wR2 = 0.1227 | wR2 = 0.1040 | wR2 = 0.1097 | |
| Extinction coefficient | 0.023(4) | n/a | n/a | 0.012(2) |
| Largest difference peak and hole (e·Å−3) | 0.177 and −0.189 | 0.181 and −0.254 | 0.202 and −0.145 | 0.245 and −0.239 |
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Habala, L.; Varényi, S.; Bilková, A.; Herich, P.; Valentová, J.; Kožíšek, J.; Devínsky, F. Antimicrobial Activity and Urease Inhibition of Schiff Bases Derived from Isoniazid and Fluorinated Benzaldehydes and of Their Copper(II) Complexes. Molecules 2016, 21, 1742. https://doi.org/10.3390/molecules21121742
Habala L, Varényi S, Bilková A, Herich P, Valentová J, Kožíšek J, Devínsky F. Antimicrobial Activity and Urease Inhibition of Schiff Bases Derived from Isoniazid and Fluorinated Benzaldehydes and of Their Copper(II) Complexes. Molecules. 2016; 21(12):1742. https://doi.org/10.3390/molecules21121742
Chicago/Turabian StyleHabala, Ladislav, Samuel Varényi, Andrea Bilková, Peter Herich, Jindra Valentová, Jozef Kožíšek, and Ferdinand Devínsky. 2016. "Antimicrobial Activity and Urease Inhibition of Schiff Bases Derived from Isoniazid and Fluorinated Benzaldehydes and of Their Copper(II) Complexes" Molecules 21, no. 12: 1742. https://doi.org/10.3390/molecules21121742
APA StyleHabala, L., Varényi, S., Bilková, A., Herich, P., Valentová, J., Kožíšek, J., & Devínsky, F. (2016). Antimicrobial Activity and Urease Inhibition of Schiff Bases Derived from Isoniazid and Fluorinated Benzaldehydes and of Their Copper(II) Complexes. Molecules, 21(12), 1742. https://doi.org/10.3390/molecules21121742