2.1. Studies on an Array of Single Stranded DNA Templates
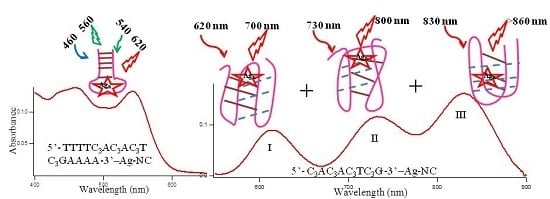
Figure 1A–C shows typical absorption spectra of Ag-NCs on seventeen oligonucleotide templates (see
Scheme 1 for sequences of oligonucleotides). The absorption of the nucleobases in the UV region are not shown here. The Ag-NCs are made in pH 6.5 citrate buffer with DNA (15 μM), AgNO
3 (90 μM) and NaBH
4 (90 μM) following the protocol as discussed below. Silver nanoparticles (NP) are formed during the process, which is evident by the absorption band ~400–420 nm [
3]. A [DNA]:[Ag] of 1:6, was chosen to optimize the Ag nanoparticle formation, while forming all the fluorescing species of the Ag-NC on a particular DNA template. Examination of the absorption spectra in
Figure 1 reveals three distinct spectral regions which were assigned to three different species of Ag-NC: species I (620–650 nm), species II (720–750 nm), and species III (~800). The population of these three species appear to be strongly dependent on the sequence of the oligonucleotides (see also
Table 1). The presence of “G” at X
iv with “T” at the X
iii positions seem to play important roles in the formation of the three species (e.g., 3T4G), while the reverse is not true (e.g., 3G4T, forms mainly species II similar to 3G, 3T, and 3T4T). The template 4G forms species I and II with a partial III. Other sequences with G at X
iv, while “T” at X
i or X
ii does not form similar NCs. When “T” or “C” is present at all X
s, the absorption spectra are drastically different. The template 3G forms Ag-NCs that are different from 4G. When a stretch of four complementary bases (T
4 and A
4) is added to the 5′ and 3′ ends of 3T4G and 3G4T to form MB3T4G (greenish-yellow solid line) and MB3G4T (light green line) in
Figure 1, the absorption spectra change drastically. It is evident from the sequences of MB3T4G and MB3G4T that the addition of the T
4 and A
4 stretches help the DNA to fold like a hairpin (
Scheme 1B), which is responsible for the dramatic differences between the absorption spectra of 3T4G and MB3T4G as well as 3G4T and MB3G4T. These results suggest that although the secondary interactions of the template are destroyed by heating at boiling temperature during the Ag-NC synthesis (see the Experimental Section below), the Ag-NC conjugated scaffold renatures back depending on its intrinsic sequence. This is further discussed below in the CD section. The size of the nanocluster and the sites of its formation dictate its optical properties.
Figure 2 depicts the fluorescence spectra of the Ag-NCs grown on the seventeen oligonucleotide templates. Depending on the absorption profiles, the NCs are excited at 730 nm (
Figure 2A, species II) and 620 nm (
Figure 2B, species I) for most of the NCs, except the ones on C
16, C
3Tim, MB3T4G, and MB3G4T templates where λ
ex = 460 nm (
Figure 2C) and 540 nm (
Figure 2D) nm.
Table 1 summarizes the fluorescence maxima (
) of all the species for the 17 templates. The
Figure 2E represents fluorescence excitation spectra monitored at emission wavelengths of 600 nm for MB3T4G (yellow), 700 nm and 800 nm for both 4G (blue) and 3T4G (red) in buffer (solid lines) and in CDx (dashed lines) environments, where observed
are ~500 nm for MB3T4G and 610, 720 nm for both 4G (blue) and 3T4G (red) templates. The observation of a small excitation band at ~610 nm for emission wavelength at 800 nm indicates the possibility of FRET [
22,
23] between species I and II. It might be happening if two clusters are forming on the same template within a few nanometers, however further studies are needed to confirm.
Figure 1 and
Figure 2 reveal that the oligonucleotides (5′-C
3X
iC
3X
iiC
3X
iiiC
3X
iv-3′) with “G” and “T” at X
iii and X
iv are important to grow species I and II which emit at ~700 and 800 nm, respectively. 3T4G and MB3T4G have drastically different fluorescence emission spectra, which supports observations from
Figure 1. These observations raised the importance of studying the secondary structures of these single stranded DNA oligonucleotides. Due to the limitation of the spectral range of the detector in the spectrofluorometer used for this study, no steady state emission studies with 830 nm excitation were performed. However, the size exclusion chromatographic measurements using the fluorescence detector allowed us to study the species III at 830 nm/860 nm (λ
ex/λ
em), which is discussed below.
Preliminary fluorescence lifetime measurements were performed with the aim of obtaining better insights regarding the distributions and microenvironments of the Ag-NCs on the C-rich DNA templates [
24].
Table 2 lists the average fluorescence lifetimes (
) along with double exponential decay components (τ
1, τ
2 ns) for species I of selected DNA templates and
Figure S1 presents the decay profiles. Although there is no difference in (
) between 3T4G, 3T and 4G in buffer, the population distribution of the minor (τ
1) and major (τ
2) components vary between 3T4G and 3T/4G scaffolds. The lifetime of the major component decreased from 2.8 (in 3T/4G ) to 2.3 ns in 3T4G which indicates that the microenvironments of the Ag-NC species I are different in the above matrices. There is a drastic difference of (
) between Ag-NCs enclosed in 3T4G/4G/3T and MB3T4G, C
16 and C
3Tim templates, implying variations in microenvironments. This is discussed using CD spectroscopic measurements which are given below. The presence of two absorption bands in
Figure 1 for MB3T4G templated Ag-NC indicated two silver nanocluster species which are supported by two very distinct (
) of 1.7 and 4.6 ns with λ
ex/λ
em = 460/535 and 496/620 nm, respectively. Two distinct lifetime components (biexponential decay), for mostly all decay studies (λ
ex/λ
em) on most of the nanoclusters, are indicative of heterogeneity in the microenvironment, [
24,
25] which is related to the various structural motifs (loops, grooves etc.) present on a DNA template (see
Scheme 1A,B). It is pertinent to mention that unlike fluorescence emission (
) which is usually environment sensitive, the observed lifetime value depends on both radiative as well as non-radiative decay parameters Equation (2) [
24], some of which are likely to depend on other factors besides polarity or hydrophobicity of the surrounding.
Circular dichroism (CD) spectra provide diagnostic signatures for the presence of secondary structures in the single stranded DNA sequence. A characteristic positive peak at ~288 nm and a negative peak at ~265 nm, with crossover at 276 nm, indicate the formation of an i-motif structure [
26,
27]. A major difference between the CD spectra of the spontaneously renatured naked DNA and Ag-NC conjugated DNA template is observable in
Figure S2, which clearly indicates the importance of Ag-NC in the refolding process.
Figure 3 presents the CD spectra of the DNA templates in free (A) and with Ag-NCs (B) at room temperature in 10 mM citrate buffer at pH 6.5. Most of the DNA sequences (see
Scheme 1A) showed presence of weak i-motifs (with negative and positive bands at ~258 and 285 nm) except oligonucleotides C
16, MB3T4G and MB3G4T which have distinct CD spectra. C
16 shows positive and negative bands at 266 and 288 nm, while MB3T4G and MB3G4T show negative bands at ~252 nm. The CD spectra of the Ag-NC conjugated DNA templates showed characteristic negative and positive bands at ~261 and 287 nm (in
Figure 3B) which indicates a classic i-motif folding of the scaffold. As is mentioned before, the DNA templates were denatured during the formation of the Ag-NCs. Hence the stronger features of i-motif (compared to
Figure 3A) in the CD spectra of the Ag-NC conjugated DNA templates at pH 6.5, are indicative of Ag induced refolding of the templates. The presence of complementary bases at the two ends of MB3T4G and MB3G4T helps in folding these oligonucleotides in a unique hairpin form (
Scheme 1B), which is responsible for the observed differences in their CD spectra compared to other templates in
Figure 3. MB3T4G has five base pairs in the hairpin stem, while MB3G4T has four, which gives rise to the distinct microenvironments of the enclosed Ag-NC on MB3T4G/MB3G4T templates, and explains the dissimilarities between themselves. According to Dettler
et al. [
26], there are at least three i-motif-like folded conformations for a single cytosine rich DNA oligonucleotide, which can co-exist in a given solution. The observed CD spectra are an ensemble of all the i-motif-like conformers at a given condition and a single base change can give rise to a different i-motif. Hence the variations in the CD spectral profiles between all the C-rich DNA (except MB3T4G and MB3G4T) are due to the presence of various i-motif-like conformers for a particular template, which is the reason for the occurrence of distinguished absorption and fluorescence emission of Ag-NC on each scaffold, as was observed in
Figure 1 and
Figure 2 and
Table 1.
2.2. Influence of pH and Increasing Silver Concentrations on the Growth of Species I, II, III
The coexistence of species I, II, and III on 3T4G makes this DNA template an ideal candidate to study the interconversion between the three species. The absorption spectra of 3T4G NC (with 15 μM DNA and 90 μM AgNO
3 and NaBH
4) with increasing pH from 5 to 7.5, are presented in
Figure 4A. With increase in pH from 5 to 5.5, absorbance of species I decreased with increase in II and III (more than II). At pH 6 and 6.5, species III has the highest absorbance, which decreases at higher pH (7 and 7.5), with a simultaneous growth in the absorbances of I and II. The increase in the growth of species III with increase in pH until pH 6.5 suggests that species III forms maximally at low H
+. This indicates that a balance between H
+ and Ag
+ is needed to form species III. Circular dichroism studies indicated that the template Ag-NC conjugated 3T4G forms a classic i-motif at pH 6.5 (
Figure 3B), which drives the formation of species III. This agrees very well with the findings of Day
et al. [
27], where Ag
+ ions induced i-motif folding has been observed at neutral and basic pH conditions. This is explained in detail using CD spectroscopic measurements as given below.
Figure 4B provides the absorption spectra of Ag-NC grown on 3T4G using different concentrations of Ag
+. It is observed that with increase in [Ag
+] from 15 (DNA:Ag = 1:1) to 120 μM (DNA:Ag = 1:8), there is an increase in growth of species I, II, and III. Species III appears to start forming at Ag
+ concentration of 60 μM and at 120 μM it has the highest absorbance. This is indicative of the association of a larger Ag cluster with species III. We studied the aging of the nanocluster species for the 1DNA: 6Ag samples of 3T4G, for five days which is shown in
Figure S3. We observed that absorbance of species II and III decreased over time with a simultaneous increase in species I. This is due to the conversion of the templates from actual i-motif to mostly i-motif-like topologies with time. The freshly prepared 3T4G enclosed Ag-NC solution has mixture of all conformers of i-motif (i-motif-like) including the classical i-motif (as discussed in the previous section on CD spectroscopy), which slowly converts to i-motif-like forms, thereby increasing the concentrations of Species I. It is pertinent to mention that species II is previously shown to be made up of 10 Ag atoms [
20].
Figure S4 provides the fluorescence emission spectra of 3T4G template Ag-NCs in different pH solution. Fluorescence intensity of species II increases until pH 7, which is shown by the red profile in
Figure 5A. For species I, there is a little increase in fluorescence intensity with increase in pH from 5.5 to 7.5. However, there is a sharp blue shift is the
of species II (as is shown
Figure 5B) from 799 (pH 5) to 787 (pH 6.5), which gradually levels off at higher pH. This could be either due to the dipolar relaxation process, indicating the less polar surrounding matrix around the excited NC fluorophore at higher pH and/or due to the change in the structure of the microenvironment. The former is discussed in the next section. To clarify the possible reason, we looked at the secondary structure of the nanocluster scaffold at different pH.
Figure 6A shows the CD spectra of Ag nanocluster conjugated 3T4G template at pH 5–7.5, where signatures of i-motif-like structures are evident in all samples. However at pH 6 and 6.5, the classic profiles of i-motif (negative and positive bands at 260 and 288 nm) could be observed, which supports our findings from
Figure 4A,B that species III needs an accurate i-motif template, which only takes place at the correct combination of Ag
+ and H
+ ion concentrations.
Figure 6B magnified the
Figure 6A spectra between 350 and 450 nm, where nanocluster induced asymmetric nature of the template can be observed. At higher pH (7, 7.5), where the i-motif features become weaker (according to
Figure 6A), the absorbance of species III decreased, which was shown in
Figure 4A. Hence species I and II are grown in the loosely folded i-motif-like structures which mainly exist at neutral to slightly basic pH.
Fluorescence emission spectra of species I and II with increasing concentrations of Ag
+, when excited at 620 (I) and 730 (II) nm, respectively, are shown in
Figure S5, where the changes are consistent with the changes in absorbance (
Figure 4B). Although no significant change in the
of species I is observed, there is a 5 nm red shift in the
of species II for the 1:8 ([DNA]:[Ag]) 3T4G nanoclusters. There are two possible reasons for this: i. An appreciable change in the microenvironment of species II at higher Ag
+ concentrations, ii. The interference of the fluorescence of species III, due to its higher absorption at higher Ag
+. In the earlier section we have observed that the growth of species III is associated with the correct folding of the template in an i-motif manner at higher Ag
+ and lower H
+ concentrations. The red shift of the
of species II at higher Ag
+ is thus significantly associated with both the change in microenvironment of the Ag-NCs and interference from species III. The influence of different salt (NaNO
3) concentrations on the formation of the 3T4G enclosed Ag-NC was studied, using absorption and CD spectroscopy, which are shown in
Figure S6. With increase in salt from 60 to 100 mM, although no change in the secondary structure of the template was observed, there was a significant decrease in the absorbance of species I, II, and III. With further increase in salt to 200 mM, an appreciable increase in the helicity of 3T4G scaffold was observed, but the absorbances were further reduced. It is noteworthy that there was no change in the absorbance of the Ag nanoparticle at ~420 nm at the increased salt conditions. These results suggest that higher concentrations of Na
+ ions prevent the formation of Ag-NC on the template which reduces the overall productions of species I, II, and III.
2.3. Influence of the Polarity of the Microenvironments on Species I, II, III
One of the motivations of undertaking the present study is to inquire whether one can observe manifestations of dipolar relaxation processes [
22,
24,
25,
28] around the excited DNA enclosed Ag-NC molecules when incorporated in the matrices of varied polarities. Solvent dipolar reorientation, which responds strongly to variations in the fluorophore environments [
24,
25], can be an important relaxation mechanism for the excited S
1 state of the Ag-NC. A convenient way to analyze dipolar relaxation process by steady state fluorescence spectroscopy is to mix the NC in a confined environment such as cyclodextrin. The use of cyclodextrin (both naturally derived, and chemically modified varieties) as “nano vehicles” for therapeutic drugs [
29,
30,
31], has become a common method in recent years due to their capabilities of enhancing solubility of hydrophobic drugs, their dissolution rate, and increased membrane permeability. With a hydrophilic outer surface and comparatively hydrophobic central cavity, cyclodextrins can form water soluble inclusion complexes with a variety of molecules. In this study, we have used methyl-beta-cyclodextrin (CDx) [
29] as well as polar protic (e.g., water, buffer, ethylene glycol) and aprotic (dioxane, DMSO) solvents to study the effect of microenvironments of varied polarities, on the optical properties of 3T4G enclosed Ag-NCs, which are displayed by the absorption and fluorescence spectra in
Figure 7. Interestingly, the absorbance maxima (
Figure 7 top) of species I blue shifts by 3 nm in CDx compared to that in water/buffer. The CD spectrum of 3T4G templated Ag-NC in the presence of CDx media (not shown) is similar to that in buffer, suggesting that the secondary structure of the DNA template is not altered by CDx [
31]. Fluorescence anisotropy (
r) [
22,
31] was determined using Equation (1). “
r” of species II was found to be 0.24 and 0.26 in buffer and CDx matrix, respectively. “
r” is a measure of the rigidity of the surrounding environment [
22]. A low value of “
r” is indicative of considerable motional freedom of the fluorophore, where “
r” = 0.4 is the theoretical maximum limit [
22,
25,
31]. The “
r” of 0.24 suggests that Ag-NC are motionally constrained inside the DNA template, and this restriction increases in CDx environment. Polar aprotic solvent DMSO only allowed the formation of species I of 3T4G Ag-NC. Dioxane destabilized the DNA template (as was evident from CD spectra, not shown) and hence the cluster formation did not take place. However, there is a 3 nm red shift in the emission maxima of species I and II in protic solvent ethylene glycol (relative polarity 0.794) [
25] with respect to buffer, which implies that 3T4G templated Ag-NC can appreciably probe the environmental variations. Hence, the shifts in
of 3T4G enclosed Ag-NCs observed in
Figure 5B can also be attributed to a difference of the dipole moments between ground (S
0) and excited (S
1) states, resulting in noticeable solvent dipolar relaxation around the chromophore [
25]. We studied the stabilities of the species I, II, and III in all the solvents for three consecutive days using absorption and fluorescence parameters.
Figure S7 shows that the intensities of species I increases at the cost of II and III. This increase is more in ethylene glycol environment and may be attributed to the viscous nature of the solvent. This opens the door to the use of viscous solvents in tuning and growing of an intended NC species.
2.4. Size Exclusion Chromatographic (SEC) Studies
Although, the importance of the secondary structures (folding) of the single stranded DNA templates on the growth of the NIR emitting Ag-NCs were emphasized by pH and Ag
+ concentration dependent studies using absorbance, fluorescence, and circular dichroism spectroscopy, the molecularity of the templates remained unknown. SEC provides an easy way to understand the folded nature along with the number of templates associated with the species I, II, III [
16].
Figure 8A provides the size exclusion chromatogram (using fluorescence) of 3T4G NC species I (λ
ex = 620/λ
em = 700 nm), II (λ
ex = 730/λ
em = 800 nm) and III (λ
ex = 830/λ
em = 860 nm). It is noteworthy that, due to the sensitivity limitation of the detector used for fluorescence measurements in the NIR region, the exact
of species III are not known. Hence, λ
em = 860 nm for detecting species III in the fluorescence detector of SEC (see Materials and Methods), is an approximation. The retention times of species III (10.89 min) > II (10.84 min) > I (10.72 min) prove that species III has the most compact structure, which agrees very well with our findings from CD spectroscopic measurements which showed that species III required a fully folded, classical i-motif to form. Single stranded thymine (T) oligonucleotides of different sizes (5, 16, 24, 30 bases long) were used to draw a standard curve, with the assumption that thymine oligonucleotides do not undergo folding [
16].
Figure 8B presents the best fit line of the plot between retention time and log of molecular mass of “T” DNAs. Using slope and intercept of the standard curve, the “observed molecular mass” of the unconjugated 3T4G was calculated to be 2630 (designated by “4” in
Figure 8B), which is around half of its actual molecular mass (4668), confirming the folded nature of the free unimolecular template. CD spectra of the unconjugated DNA at pH 6.5 (
Figure 3A) showed weak i-motif folding, which is confirmed by SEC. Similarly, the “observed molecular mass” of MB3T4G templated Ag-NC (using fluorescence with λ
ex = 540 nm) was found to be 2209, compared to the actual mass of 7138 g/mole, which is consistent with the hairpin like (
Scheme 1B) folded structure for MB3T4G. The mass of the species I, II, III (
Figure 8A) were calculated to be 2948, 2531, 2366 (shown by “5”, “3”, “2” in
Figure 8B). The molecular mass of CDx bound species I of 3T4G-NC (“6” in
Figure 8B) was found to be 3936, which proves that one cyclodextrin molecule conjugates with the Ag-NC species (the average molecular mass of CDx is ~1310). Species I and II must be associated with i-motif-like topologies. SEC measurements confirmed the uni-intramolecular folding nature of the templates and supported the findings from CD spectroscopic studies.