The Pleiotropic Antibacterial Mechanisms of Ursolic Acid against Methicillin-Resistant Staphylococcus aureus (MRSA)
Abstract
:1. Introduction
2. Results
2.1. Effects of Antibiotics and Ursolic Acid on Bacterial and Mammalian Membrane Viability
2.2. Effects of Ursolic Acid on Bacterial Protein Synthesis
3. Discussion
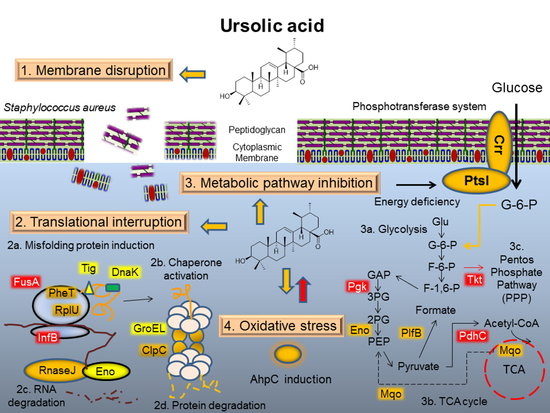
3.1. Mode of Action I: Membrane Disruption
3.2. Mode of Action II: Translation Interruption
3.3. Mode of Action III: Metabolic Pathway Interaction
3.4. Mode of Action IV: Oxidative Stress Response
4. Materials and Methods
4.1. Effects of Antimicrobial Agents on Bacterial and Mammalian Membranes Viability
4.2. 2D Gel Electrophoresis Analysis
4.3. In-Gel-Preparation of Tryptic Peptides
4.4. LC-MS/MS Analysis
4.5. Data Analysis
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shanmugam, M.K.; Nguyen, A.H.; Kumar, A.P.; Tan, B.K.; Sethi, G. Targeted inhibition of tumor proliferation, survival, and metastasis by pentacyclic triterpenoids: Potential role in prevention and therapy of cancer. Cancer Lett. 2012, 320, 158–170. [Google Scholar] [CrossRef] [PubMed]
- Way, T.D.; Tsai, S.J.; Wang, C.M.; Ho, C.T.; Chou, C.H. Chemical constituents of Rhododendron formosanum show pronounced growth inhibitory effect on non-small-cell lung carcinoma cells. J. Agric. Food Chem. 2014, 62, 875–884. [Google Scholar] [CrossRef] [PubMed]
- Baliga, M.S. Review of the phytochemical, pharmacological and toxicological properties of Alstonia scholaris Linn. R. Br (Saptaparna). Chin. J. Integr. Med. 2012, 18. [Google Scholar] [CrossRef] [PubMed]
- El-Askary, H.I.; El-Olemy, M.M.; Salama, M.M.; Sleem, A.A.; Amer, M.H. Bioguided isolation of pentacyclic triterpenes from the leaves of Alstonia scholaris (Linn.) R. Br. growing in Egypt. Nat. Prod. Res. 2012, 26, 1755–1758. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.-M.; Chen, H.-T.; Wu, Z.-Y.; Jhan, Y.-L.; Shyu, C.-L.; Chou, C.-H. Antibacterial and synergistic activity of pentacyclic triterpenoids isolated from Alstonia scholaris. Molecules 2016, 21, 139. [Google Scholar] [CrossRef] [PubMed]
- de Leon, L.; Beltran, B.; Moujir, L. Antimicrobial activity of 6-oxophenolic triterpenoids. Mode of action against Bacillus subtilis. Planta Med. 2005, 71, 313–319. [Google Scholar] [CrossRef] [PubMed]
- Katerere, D.R.; Gray, A.I.; Nash, R.J.; Waigh, R.D. Antimicrobial activity of pentacyclic triterpenes isolated from African Combretaceae. Phytochemistry 2003, 63, 81–88. [Google Scholar] [CrossRef]
- Hecker, M.; Reder, A.; Fuchs, S.; Pagels, M.; Engelmann, S. Physiological proteomics and stress/starvation responses in Bacillus subtilis and Staphylococcus aureus. Res. Microbiol. 2009, 160, 245–258. [Google Scholar] [CrossRef] [PubMed]
- Bandow, J.E.; Brotz, H.; Leichert, L.I.; Labischinski, H.; Hecker, M. Proteomic approach to understanding antibiotic action. Antimicrob. Agents Chemother. 2003, 47, 948–955. [Google Scholar] [CrossRef] [PubMed]
- Evers, S.; Di Padova, K.; Meyer, M.; Langen, H.; Fountoulakis, M.; Keck, W.; Gray, C.P. Mechanism-related changes in the gene transcription and protein synthesis patterns of Haemophilus influenzae after treatment with transcriptional and translational inhibitors. Proteomics 2001, 1, 522–544. [Google Scholar] [CrossRef]
- Dennis, G., Jr.; Sherman, B.T.; Hosack, D.A.; Yang, J.; Gao, W.; Lane, H.C.; Lempicki, R.A. DAVID: Database for annotation, visualization, and integrated discovery. Genome Biol. 2003, 4, P3. [Google Scholar] [CrossRef]
- Szklarczyk, D.; Franceschini, A.; Wyder, S.; Forslund, K.; Heller, D.; Huerta-Cepas, J.; Simonovic, M.; Roth, A.; Santos, A.; Tsafou, K.P.; et al. STRING v10: Protein-protein interaction networks, integrated over the tree of life. Nucleic Acids Res. 2015, 43, D447–D452. [Google Scholar] [CrossRef] [PubMed]
- Kurek, A.; Nadkowska, P.; Pliszka, S.; Wolska, K.I. Modulation of antibiotic resistance in bacterial pathogens by oleanolic acid and ursolic acid. Phytomedicine 2012, 19, 515–519. [Google Scholar] [CrossRef] [PubMed]
- Kuroda, M.; Nagasaki, S.; Ohta, T. Sesquiterpene farnesol inhibits recycling of the C55 lipid carrier of the murein monomer precursor contributing to increased susceptibility to beta-lactams in methicillin-resistant Staphylococcus aureus. J. Antimicrob. Chemother. 2007, 59, 425–432. [Google Scholar] [CrossRef] [PubMed]
- Hurdle, J.G.; O’Neill, A.J.; Chopra, I.; Lee, R.E. Targeting bacterial membrane function: An underexploited mechanism for treating persistent infections. Nat. Rev. Microbiol. 2011, 9, 62–75. [Google Scholar] [CrossRef] [PubMed]
- Ooi, N.; Miller, K.; Hobbs, J.; Rhys-Williams, W.; Love, W.; Chopra, I. XF-73, a novel antistaphylococcal membrane-active agent with rapid bactericidal activity. J. Antimicrob. Chemother. 2009, 64, 735–740. [Google Scholar] [CrossRef] [PubMed]
- Chatterjee, I.; Becker, P.; Grundmeier, M.; Bischoff, M.; Somerville, G.A.; Peters, G.; Sinha, B.; Harraghy, N.; Proctor, R.A.; Herrmann, M. Staphylococcus aureus ClpC is required for stress resistance, aconitase activity, growth recovery, and death. J. Bacteriol. 2005, 187, 4488–4496. [Google Scholar] [CrossRef] [PubMed]
- Michel, A.; Agerer, F.; Hauck, C.R.; Herrmann, M.; Ullrich, J.; Hacker, J.; Ohlsen, K. Global regulatory impact of ClpP protease of Staphylococcus aureus on regulons involved in virulence, oxidative stress response, autolysis, and DNA repair. J. Bacteriol. 2006, 188, 5783–5796. [Google Scholar] [CrossRef] [PubMed]
- Frees, D.; Chastanet, A.; Qazi, S.; Sorensen, K.; Hill, P.; Msadek, T.; Ingmer, H. Clp ATPases are required for stress tolerance, intracellular replication and biofilm formation in Staphylococcus aureus. Mol. Microbiol. 2004, 54, 1445–1462. [Google Scholar] [CrossRef] [PubMed]
- Flamholz, A.; Noor, E.; Bar-Even, A.; Liebermeister, W.; Milo, R. Glycolytic strategy as a tradeoff between energy yield and protein cost. Proc. Natl. Acad. Sci. USA 2013, 110, 10039–10044. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.W.; Chung, C.H.; Ma, T.Y.; Wong, H.C. Roles of alkyl hydroperoxide reductase subunit C (AhpC) in viable but nonculturable Vibrio parahaemolyticus. Appl. Environ. Microbiol. 2013, 79, 3734–3743. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Xie, Q.W.; Nathan, C. Alkyl hydroperoxide reductase subunit C (AhpC) protects bacterial and human cells against reactive nitrogen intermediates. Mol. Cell 1998, 1, 795–805. [Google Scholar] [CrossRef]
- Kohanski, M.A.; Dwyer, D.J.; Hayete, B.; Lawrence, C.A.; Collins, J.J. A common mechanism of cellular death induced by bactericidal antibiotics. Cell 2007, 130, 797–810. [Google Scholar] [CrossRef] [PubMed]
- Oliva, B.; Miller, K.; Caggiano, N.; O’Neill, A.J.; Cuny, G.D.; Hoemarm, M.Z.; Hauske, J.R.; Chopra, I. Biological properties of novel antistaphylococcal quinoline-indole agents. Antimicrob. Agents Chemother. 2003, 47, 458–466. [Google Scholar] [CrossRef] [PubMed]






| Antimicrobial Compounds | Bacterial Membrane Integrity (%) | Erythrocyte Integrity (%) |
|---|---|---|
| None | 99.4 ± 7.5 a | 101.8 ± 2.9 a |
| 5% SDS | 0 ± 0 d | 0 ± 0 c |
| Ampicillin | 17.9 ± 0.6 c | 81.0 ± 2.6 b |
| Tetracycline | 87.3 ± 9.6 a | 82.2 ± 2.3 b |
| Ursolic acid | 49.5 ± 0.8 b | 83.0 ± 5.0 b |
| Protein | Protein Name | Accession No. | pI | M.W. | Coverage (%) |
|---|---|---|---|---|---|
| Adh | Alcohol dehydrogenase | gi|487362910 | 5.63 | 94,886 | 30% |
| AhpC | Alkyl hydroperoxide reductase subunit C | gi|445974926 | 5.06 | 20,846 | 38% |
| AKRs | glyoxal reductase | gi|446374225 | 5.09 | 31,261 | 14% |
| AlaS | Alanyl-tRNA synthase | gi|446656721 | 5.00 | 98,479 | 13% |
| Asp23 | alkaline shock protein 23 | gi|446137381 | 5.27 | 19,060 | 27% |
| AtpD | ATP synthase subunit beta | gi|446433275 | 4.71 | 51,382 | 40% |
| ClpC | ATP-dependent Clp protease, ATP-binding subunit ClpC | gi|446819870 | 5.51 | 90,968 | 11% |
| Crr | Glucose-specific phosphotransferase enzyme IIA | gi|261278560 | 4.64 | 63,097 | 15% |
| DnaK | Chaperone protein DnaK | gi|445956852 | 4.70 | 66,319 | 40% |
| Eno | Enolase | gi|447044501 | 4.58 | 47,115 | 43% |
| FusA | Translation elongation factor G | gi|395759323 | 4.80 | 76,530 | 37% |
| GroEL | Chaperonin protein, 60 kDa | gi|657020658 | 4.59 | 57,537 | 16% |
| InfB | Translation initiation factor IF-2 | gi|445965771 | 5.09 | 77,795 | 18% |
| Mqo2 | Malate quinone oxidoreductase 2 | gi|447052792 | 6.12 | 55,978 | 13% |
| PdhC | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | gi|2499415 | 4.87 | 46,411 | 17% |
| PflB | Formate acetyltransferase | gi|446817402 | 5.31 | 84,822 | 24% |
| Pgk | Phosphoglycerate kinase | gi|446997500 | 5.17 | 42,603 | 42% |
| PheT | phenylalanyl-tRNA synthase subunit beta | gi|446831715 | 4.71 | 88,838 | 19% |
| Prs | Ribose-phosphate pyrophosphokinase | gi|446856516 | 5.88 | 35,292 | 11% |
| PtsI | Phosphoenolpyruvate protein phosphotransferase | gi|446696933 | 4.82 | 34,929 | 11% |
| Pyk | Pyruvate kinase | gi|447155392 | 5.23 | 63,063 | 47% |
| RnaseJ | Ribonuclease J2 | gi|445974731 | 5.81 | 62,591 | 18% |
| RplU | 50S ribosomal protein L21 | gi|75530481 | 9.78 | 11,309 | 50% |
| RpoA | RNA polymerase, α chain | gi|686416814 | 4.66 | 34,947 | 24% |
| RpoB | RNA polymerase, β chain | gi|686122810 | 4.91 | 133,152 | 19% |
| SufB | Fe-S cluster assembly protein | gi|446997144 | 5.08 | 52,498 | 19% |
| Tig | Trigger factor | gi|446049710 | 4.34 | 48,577 | 27% |
| Tkt | Transketolase | gi|446403587 | 5.00 | 72,212 | 34% |
| TufA | Translation elongation factor Tu | gi|446963310 | 4.77 | 43,077 | 56% |
| Protein | Protein Name | Ratio 1 Treatment/Control |
|---|---|---|
| Transcription | ||
| RpoB | RNA polymerase, β chain | 1.042 |
| RpoA | RNA polymerase, α chain | 1.123 |
| Translation | ||
| FusA | Translation elongation factor G | 0.685 |
| TufA | Translation elongation factor Tu | 0.958 |
| InfB | Translation initiation factor IF-2 | 0.332 |
| AlaS | Alanyl-tRNA synthase | 0.975 |
| PheT | phenylalanyl-tRNA synthase subunit beta | 2.676 |
| RplU | 50S ribosomal protein L21 | 8.798 |
| Protein folding and RNA degradation | ||
| ClpC | ATP-dependent Clp protease, subunit ClpC | 5.602 |
| GroEL | Chaperonin protein, 60 kDa | 1.387 |
| Tig | Trigger factor | 1.368 |
| DnaK | Chaperone protein DnaK | 1.282 |
| Eno | Enolase | 1.387 |
| RnaseJ | Ribonuclease J2 | 2.091 |
| Glycolysis, TCA cycle and Pentose phosphate pathway | ||
| PflB | Formate acetyltransferase | 1.978 |
| Pgk | Phosphoglycerate kinase | 0.386 |
| PdhC | Dihydrolipoyllysine-residue acetyltransferase component of pyruvate dehydrogenase complex | 0.662 |
| Pyk | Pyruvate kinase | 1.118 |
| Tkt | Transketolase | 0.577 |
| Mqo2 | Malate quinone oxidoreductase 2 | 4.11 |
| Phosphotransferase system | ||
| PtsI | Phosphoenolpyruvate-protein phosphotransferase | 2.32 |
| Crr | Glucose-specific phosphotransferase enzyme IIA | 1.671 |
| Oxidative stress | ||
| AhpC | Alkyl hydroperoxide reductase subunit C | 6.074 |
| Others | ||
| Adh | Alcohol dehydrogenase | 4.213 |
| SufB | Fe-S cluster assembly protein | 0.235 |
| AKRs | Glyoxal reductase | 0.708 |
| Prs | Ribose-phosphate pyrophosphokinase | 0.869 |
| AtpD | ATP synthase subunit beta | 1.473 |
| Asp23 | alkaline shock protein 23 | 0.265 |
- Sample Availability: Samples of the compounds are available from the authors.
© 2016 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license ( http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, C.-M.; Jhan, Y.-L.; Tsai, S.-J.; Chou, C.-H. The Pleiotropic Antibacterial Mechanisms of Ursolic Acid against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules 2016, 21, 884. https://doi.org/10.3390/molecules21070884
Wang C-M, Jhan Y-L, Tsai S-J, Chou C-H. The Pleiotropic Antibacterial Mechanisms of Ursolic Acid against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules. 2016; 21(7):884. https://doi.org/10.3390/molecules21070884
Chicago/Turabian StyleWang, Chao-Min, Yun-Lian Jhan, Shang-Jie Tsai, and Chang-Hung Chou. 2016. "The Pleiotropic Antibacterial Mechanisms of Ursolic Acid against Methicillin-Resistant Staphylococcus aureus (MRSA)" Molecules 21, no. 7: 884. https://doi.org/10.3390/molecules21070884
APA StyleWang, C. -M., Jhan, Y. -L., Tsai, S. -J., & Chou, C. -H. (2016). The Pleiotropic Antibacterial Mechanisms of Ursolic Acid against Methicillin-Resistant Staphylococcus aureus (MRSA). Molecules, 21(7), 884. https://doi.org/10.3390/molecules21070884