Comprehensive Characterization for Ginsenosides Biosynthesis in Ginseng Root by Integration Analysis of Chemical and Transcriptome
Abstract
:1. Introduction
2. Results
2.1. Localization of Ginsenosides in the Main Root of P. ginseng
2.2. De Novo Assembly of the P. ginseng Transcriptome
2.3. Functional Analysis of Unigenes
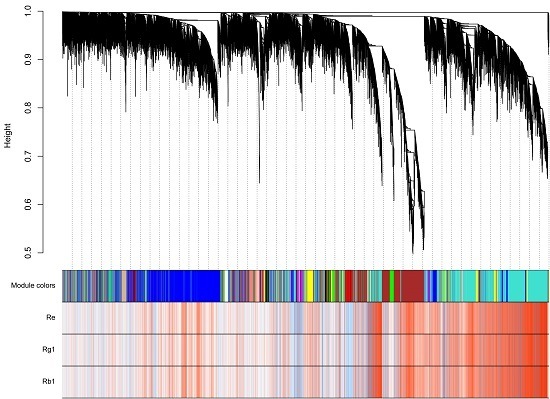
2.4. Distinct Transcriptome in Ginseng Main Root Tissues
2.5. Analysis of Triterpenoid Saponins Metabolism
3. Discussion
4. Materials and Methods
4.1. Plant Material
4.2. HPLC Analysis
4.3. RNA-Seq
4.4. Transcriptome Analysis
4.5. Data Availability
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
Abbreviations
| UGTs | UDP-glycosyltransferases |
| DEGs | Differential expression genes |
| MALDI-MS | Matrix-assisted laser desorption/ionization mass spectrometry |
| HPLC | High-performance liquid chromatography |
| NCBI Nr | NCBI non-redundant protein non-redundant protein |
| GO | Gene Ontology |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| ORF | Open read frame |
| FPKM | Fragments per kilobase of exon model per million mapped reads |
| WGCNA | Weighted gene co-expression network analysis |
| MVA | Mevalonate |
| MEP | Methylerythritol phosphate |
| FPS | Farnesyl diphosphate synthase |
| SE | Squalene epoxidase |
| Beta-ASs | Beta-amyrin synthases |
| OASs | Oleanolic acid synthases |
| DDSs | Dammarendiol synthases |
| PPDSs | Protopanaxadiol synthase |
| PPTSs | Protopanaxatriol synthases |
| AACT | Acetyl-CoA C-acetyltransferase |
| HMGS | 3-Hydroxy-3-methylglutaryl-CoA synthase |
| IspD | Isoprenoid synthase-containing protein D |
| IDI | Isopentenyl diphosphate isomerase |
| DXS | 1-Deoxy-d-xylulose-5-phosphate synthase |
| IspH | Isoprenoid synthase-containing protein H |
| DXR | 1-Deoxy-d-xylulose-5-phosphate reductoisomerase |
| IspF | Isoprenoid synthase-containing protein F |
| IspG/gcpE | Isoprenoid synthase-containing protein gcpE |
| IspE | Isoprenoid synthase-containing protein E |
References
- Block, K.I.; Mead, M.N. Immune system effects of echinacea, ginseng, and astragalus: A review. Integr. Cancer Ther. 2003, 2, 247–267. [Google Scholar] [CrossRef] [PubMed]
- Christensen, L.P. Chapter 1 Ginsenosides: Chemistry, Biosynthesis, Analysis, and Potential Health Effects. Adv. Food Nutr. Res. 2008, 55, 1–99. [Google Scholar]
- Ko, S.R.; Choi, K.J.; Han, K.W. Comparison of proximate composition, mineral nutrient, amino acid and free sugar contents of several Panax species. Korean J. Ginseng Sci. 1996, 20, 36–41. [Google Scholar]
- Attele, A.S.; Wu, J.A.; Yuan, C.S. Ginseng pharmacology: Multiple constituents and multiple actions. Biochem. Pharmacol. 1999, 58, 1685–1693. [Google Scholar] [CrossRef]
- Zhuravlev, Y.N.; Koren, O.G.; Reunova, G.D.; Muzarok, T.I.; Gorpenchenko, T.Y.; Yuliya, I.L.; Khrolenko, A. Panax ginseng natural populations: Their past, current state and perspectives. In Proceedings of the 2008 International Conference on Ginseng, Changchun, China, 1–3 September 2008; pp. 1127–1136. [Google Scholar]
- Gao, Y.; He, X.; Wu, B.; Long, Q.; Shao, T.; Wang, Z.; Wei, J.; Li, Y.; Ding, W. Time-course transcriptome analysis reveals resistance genes of Panax ginseng induced by Cylindrocarpon destructans infection using RNA-seq. PLoS ONE 2016, 11, e0149408. [Google Scholar] [CrossRef] [PubMed]
- Rahman, M.; Punja, Z.K. Factors influencing development of root rot on ginseng caused by Cylindrocarpon destructans. Phytopathology 2005, 95, 1381–1390. [Google Scholar] [CrossRef] [PubMed]
- Lü, J.M.; Yao, Q.; Chen, C.Y. Ginseng compounds: An update on their molecular mechanisms and medical applications. Curr. Vasc. Pharmacol. 2009, 7, 293–302. [Google Scholar] [CrossRef] [PubMed]
- Ruan, C.C.; Liu, Z.; Li, X.; Liu, X.; Wang, L.J.; Pan, H.Y.; Zheng, Y.N.; Sun, G.Z.; Zhang, Y.S.; Zhang, L.X. Isolation and characterization of a new ginsenoside from the fresh root of Panax ginseng. Molecules 2010, 15, 2319–2325. [Google Scholar] [CrossRef] [PubMed]
- Shi, W.; Wang, Y.; Li, J.; Zhang, H.; Ding, L. Investigation of ginsenosides in different parts and ages of Panax ginseng. Food Chem. 2007, 102, 664–668. [Google Scholar] [CrossRef]
- Bulgakov, V.P.; Khodakovskaya, M.V.; Labetskaya, N.V.; Chernoded, G.K.; Zhuravlev, Y.N. The impact of plant rolC oncogene on ginsenoside production by ginseng hairy root cultures. Phytochemistry 1998, 49, 1929–1934. [Google Scholar] [CrossRef]
- Palazón, J.; Cusidó, R.M.; Bonfill, M.; Mallol, A.; Moyano, E.; Morales, C.; Piñol, M.T. Elicitation of different Panax ginseng transformed root phenotypes for an improved ginsenoside production. Plant Physiol. Biochem. 2003, 41, 1019–1025. [Google Scholar] [CrossRef]
- Tansakul, P.; Shibuya, M.T.; Ebizuka, Y. Dammarenediol-II synthase, the first dedicated enzyme for ginsenoside biosynthesis, in Panax ginseng. FEBS Lett. 2006, 580, 5143–5149. [Google Scholar] [CrossRef] [PubMed]
- Mao, Q.; Bai, M.; Xu, J.D.; Kong, M.; Zhu, L.Y.; Zhu, H.; Wang, Q.; Li, S.L. Discrimination of leaves of Panax ginseng and P. quinquefolius by ultra high performance liquid chromatography quadrupole/time-of-flight mass spectrometry based metabolomics approach. J. Pharm. Biomed. Anal. 2014, 97C, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Xie, G.; Plumb, R.; Su, M.; Xu, Z.; Zhao, A.; Qiu, M.; Long, X.; Liu, Z.; Jia, W. Ultra-performance LC/TOF MS analysis of medicinal Panax herbs for metabolomic research. J. Sep. Sci. 2008, 31, 1015–1026. [Google Scholar] [CrossRef] [PubMed]
- Ahn, I.O.; Lee, S.S.; Lee, J.H.; Lee, M.J.; Jo, B.G. Comparison of ginsenoside contents and pattern similarity between root parts of new cultivars in Panax ginseng C.A. Meyer. In Life and Education in Early Societies; Macmillan Co.: New York, NY, USA, 2008; pp. 142–151. [Google Scholar]
- Ji, Y.O.; Kim, Y.J.; Jang, M.G.; Joo, S.C.; Kwon, W.S.; Kim, S.Y.; Jung, S.K.; Yang, D.C. Investigation of ginsenosides in different tissues after elicitor treatment in Panax ginseng. J. Ginseng Res. 2014, 38, 270–277. [Google Scholar]
- Zhang, Y.C.; Li, G.; Jiang, C.; Yang, B.; Yang, H.J.; Xu, H.Y.; Huang, L.Q. Tissue-specific distribution of ginsenosides in different aged ginseng and antioxidant activity of ginseng leaf. Molecules 2014, 19, 17381–17399. [Google Scholar] [CrossRef] [PubMed]
- Kim, N.; Kim, K.; Choi, B.Y.; Lee, D.; Shin, Y.S.; Bang, K.H.; Cha, S.W.; Lee, J.W.; Choi, H.K.; Jang, D.S.; et al. Metabolomic approach for age discrimination of Panax ginseng using UPLC-Q-Tof MS. J. Agric. Food Chem. 2011, 59, 10435–10441. [Google Scholar] [CrossRef] [PubMed]
- Shan, S.M.; Luo, J.G.; Huang, F.; Kong, L.Y. Chemical characteristics combined with bioactivity for comprehensive evaluation of Panax ginseng C.A. Meyer in different ages and seasons based on HPLC-DAD and chemometric methods. J. Pharm. Biomed. Anal. 2014, 89, 76–82. [Google Scholar] [CrossRef] [PubMed]
- Shin, Y.S.; Bang, K.H.; In, D.S.; Kim, O.T.; Hyun, D.Y.; Ahn, I.O.; Ku, B.C.; Kim, S.W.; Seong, N.S.; Cha, S.W.; et al. Fingerprinting analysis of fresh ginseng roots of different ages using 1H-NMR spectroscopy and principal components analysis. Arch. Pharm. Res. 2007, 30, 1625–1628. [Google Scholar] [CrossRef] [PubMed]
- Yang, S.O.; Shin, Y.S.; Hyun, S.H.; Cho, S.; Bang, K.H.; Lee, D.; Choi, S.P.; Choi, H.K. NMR-based metabolic profiling and differentiation of ginseng roots according to cultivation ages. J. Pharm. Biomed. Anal. 2012, 58, 19–26. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Pan, J.Y.; Xiao, X.Y.; Lin, R.C.; Cheng, Y.Y. Simultaneous determination of ginsenosides in Panax ginseng with different growth ages using high-performance liquid chromatography–mass spectrometry. Phytochem. Anal. 2006, 17, 424–430. [Google Scholar] [CrossRef] [PubMed]
- Li, C.Y.; Lau, D.T.; Dong, T.T.; Zhang, J.; Choi, R.C.; Wu, H.Q.; Wang, L.Y.; Hong, R.S.; Li, S.H.; Song, X.; et al. Dual-index evaluation of character changes in Panax ginseng C.A. Mey stored in different conditions. J. Agric. Food Chem. 2013, 61, 6568–6573. [Google Scholar] [CrossRef] [PubMed]
- Xie, Y.Y.; Luo, D.; Cheng, Y.J.; Ma, J.F.; Wang, Y.M.; Liang, Q.L.; Luo, G.A. Steaming-induced chemical transformations and holistic quality assessment of red ginseng derived from Panax ginseng by means of HPLC-ESI-MS/MSn-based multicomponent quantification fingerprint. J. Agric. Food Chem. 2012, 60, 8213–8224. [Google Scholar] [CrossRef] [PubMed]
- Bai, H.; Wang, S.; Liu, J.; Gao, D.; Jiang, Y.; Liu, H.; Cai, Z. Localization of ginsenosides in Panax ginseng with different age by matrix-assisted laser-desorption/ionization time-of-flight mass spectrometry imaging. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2015, 1026, 263–271. [Google Scholar] [CrossRef] [PubMed]
- Kubo, M.; Tani, T.; Katsuki, T.; Ishizaki, K.; Arichi, S.; Histochemistry, I. Ginsenosides in ginseng (Panax ginseng C.A. Meyer, root). J. Nat. Prod. 1980, 43, 278–284. [Google Scholar] [CrossRef]
- Liang, Z.; Chen, Y.; Liang, X.; Qin, M.; Tao, Y.; Chen, H.; Zhao, Z. Localization of ginsenosides in the rhizome and root of Panax ginseng by laser microdissection and liquid chromatography-quadrupole/time of flight-mass spectrometry. J. Pharm. Biomed. Anal. 2015, 105, 121–133. [Google Scholar] [CrossRef] [PubMed]
- Taira, S.; Ikeda, R.; Yokota, N.; Osaka, I.; Sakamoto, M.; Kato, M.; Sahashi, Y. Mass spectrometric imaging of ginsenosides localization in Panax ginseng root. Am. J. Chin. Med. 2012, 38, 485–493. [Google Scholar] [CrossRef] [PubMed]
- Han, J.Y.; In, J.G.; Kwon, Y.S.; Choi, Y.E. Regulation of ginsenoside and phytosterol biosynthesis by RNA interferences of squalene epoxidase gene in Panax ginseng. Phytochemistry 2009, 71, 36–46. [Google Scholar] [CrossRef] [PubMed]
- Kim, T.D.; Han, J.Y.; Huh, G.H.; Choi, Y.E. Expression and functional characterization of three squalene synthase genes associated with saponin biosynthesis in Panax ginseng. Plant Cell Physiol. 2011, 52, 125–137. [Google Scholar] [CrossRef] [PubMed]
- Karwah, L.; SzeTsai, W. Pharmacology of ginsenosides: A literature review. Chin. Med. 2010, 5, 20. [Google Scholar] [CrossRef]
- Mallavadhani, U.V.; Mahapatra, A.; Raja, S.S.; Manjula, C. Antifeedant activity of some pentacyclic triterpene acids and their fatty acid ester analogues. J. Agric. Food Chem. 2003, 51, 1952–1955. [Google Scholar] [CrossRef] [PubMed]
- Osbourn, A.E. Preformed antimicrobial compounds and plant defense against fungal attack. Plant Cell 1996, 8, 1821–1831. [Google Scholar] [CrossRef] [PubMed]
- Cao, H.; Nuruzzaman, M.; Xiu, H.; Huang, J.; Wu, K.; Chen, X.; Li, J.; Wang, L.; Jeong, J.H.; Park, S.J.; et al. Transcriptome analysis of methyl jasmonate-elicited Panax ginseng adventitious roots to discover putative ginsenoside biosynthesis and transport genes. Int. J. Mol. Sci. 2015, 16, 3035–3057. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhu, Y.; Xu, G.; Chao, S.; Luo, H.; Song, J.; Ying, L.; Wang, L.; Qian, J.; Chen, S. Transcriptome analysis reveals ginsenosides biosynthetic genes, microRNAs and simple sequence repeats in Panax ginseng C.A. Meyer. BMC Genom. 2013, 14, 204–205. [Google Scholar] [CrossRef] [PubMed]
- Wang, K.; Jiang, S.; Sun, C.; Lin, Y.; Yin, R.; Wang, Y.; Zhang, M. The spatial and temporal transcriptomic landscapes of ginseng, Panax ginseng C.A. Meyer. Sci. Rep. 2015, 5, 18283. [Google Scholar] [CrossRef] [PubMed]
- Jayakodi, M.; Lee, S.C.; Yun, S.L.; Park, H.S.; Kim, N.H.; Jang, W.; Lee, H.O.; Joh, H.J.; Yang, T.J. Comprehensive analysis of Panax ginseng root transcriptomes. BMC Plant Biol. 2015, 15, 1–12. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Luo, H.; Li, Y.; Sun, Y.; Wu, Q.; Niu, Y.; Song, J.; Lv, A.; Zhu, Y.; Sun, C.; et al. 454 EST analysis detects genes putatively involved in ginsenoside biosynthesis in Panax ginseng. Plant Cell Rep. 2011, 30, 1593–1601. [Google Scholar] [CrossRef] [PubMed]
- Wu, B.; Wang, M.; Ma, Y.; Yuan, L.; Lu, S. High-throughput sequencing and characterization of the small RNA transcriptome reveal features of novel and conserved microRNAs in Panax ginseng. PLoS ONE 2012, 7, e44385. [Google Scholar] [CrossRef] [PubMed]
- Breschi, A.; Djebali, S.; Gillis, J.; Pervouchine, D.D.; Dobin, A.; Davis, C.A.; Gingeras, T.R.; Guigó, R. Gene-specific patterns of expression variation across organs and species. Genome Biol. 2016, 17, 151. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.L.; Song, J.Y.; Sun, C.; Xu, J.; Zhu, Y.J.; Verpoorte, R.; Fan, T.P. Herbal genomics: Examining the biology of traditional medicines. Science 2015, 347, S27–S29. [Google Scholar]
- Chen, S.; Hui, Y.; Han, J.; Chang, L.; Song, J.; Shi, L.; Zhu, Y.; Ma, X.; Gao, T.; Pang, X. Validation of the ITS2 region as a novel DNA barcode for identifying medicinal plant species. PLoS ONE 2010, 5, e8613. [Google Scholar] [CrossRef] [PubMed]
- Chen, S.; Xu, J.; Liu, C.; Zhu, Y.; Nelson, D.R.; Zhou, S.; Li, C.; Wang, L.; Guo, X.; Sun, Y.; et al. Genome sequence of the model medicinal mushroom Ganoderma lucidum. Nat. Commun. 2012, 3, 913. [Google Scholar] [CrossRef] [PubMed]
- Xu, H.; Song, J.; Luo, H.; Zhang, Y.; Li, Q.; Zhu, Y.; Xu, J.; Li, Y.; Song, C.; Wang, B.; et al. Analysis of the genome sequence of the medicinal plant Salvia miltiorrhiza. Mol. Plant 2016, 9, 949–952. [Google Scholar] [CrossRef] [PubMed]
- Xu, Z.; Peters, R.J.; Weirather, J.; Luo, H.; Liao, B.; Zhang, X.; Zhu, Y.; Ji, A.; Zhang, B.; Hu, S.; et al. Full-length transcriptome sequences and splice variants obtained by a combination of sequencing platforms applied to different root tissues of Salvia miltiorrhiza and tanshinone biosynthesis. Plant J. Cell Mol. Biol. 2015, 82, 951. [Google Scholar] [CrossRef] [PubMed]
- Zhihai, H.; Jiang, X.; Shuiming, X.; Baosheng, L. Comparative optical genome analysis of two pangolin species: Manis pentadactyla and Manis javanica. GigaScience 2016, 5, 5. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Y.; Xu, J.; Sun, C.; Zhou, S.; Xu, H.; Nelson, D.R.; Qian, J.; Song, J.; Luo, H.; Li, X.; et al. Chromosome-level genome map provides insights into diverse defense mechanisms in the medicinal fungus Ganoderma sinense. Sci. Rep. 2015, 5, 11087. [Google Scholar] [CrossRef] [PubMed]
- Guan, R.; Zhao, Y.; Zhang, H.; Fan, G.; Liu, X.; Zhou, W.; Shi, C.; Wang, J.; Liu, W.; Liang, X.; et al. Draft genome of the living fossil Ginkgo biloba. Gigascience 2016, 5, 49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, G.; Tian, Y.; Zhang, J.; Shu, L.; Yang, S.; Wang, W.; Sheng, J.; Dong, Y.; Chen, W. Hybrid de novo genome assembly of the Chinese herbal plant danshen (Salvia miltiorrhiza Bunge). GigaScience 2015, 4, 62. [Google Scholar] [CrossRef] [PubMed]
- Liu, S.; Wang, S.; Liu, M.; Yang, F.; Zhang, H.; Liu, S.; Wang, Q.; Zhao, Y. De novo sequencing and analysis of the transcriptome of Panax ginseng in the leaf-expansion period. Mol. Med. Rep. 2016, 14, 1404. [Google Scholar] [CrossRef] [PubMed]
- Kim, Y.J.; Yang, D.C. Functional analysis of 3-hydroxy-3-methylglutaryl coenzyme a reductase encoding genes in triterpene saponin-producing ginseng. Plant Physiol. 2014, 165, 373. [Google Scholar] [CrossRef] [PubMed]
- Chan, E.C.; Yap, S.L.; Lau, A.J.; Leow, P.C.; Toh, D.F.; Koh, H.L. Ultra-performance liquid chromatography/time-of-flight mass spectrometry based metabolomics of raw and steamed Panax notoginseng. Rapid Commun. Mass Spectrom. 2007, 21, 519–528. [Google Scholar] [CrossRef] [PubMed]
- Grabherr, M.G.; Haas, B.J.; Yassour, M.; Levin, J.Z.; Thompson, D.A.; Amit, I.; Xian, A.; Fan, L.; Raychowdhury, R.; Zeng, Q.; et al. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Lieber, M. De novo transcript sequence reconstruction from RNA-seq: Reference generation and analysis with Trinity. Nat. Protoc. 2013, 8, 1494–1512. [Google Scholar]
- Trapnell, C.; Hendrickson, D.G.; Sauvageau, M.; Goff, L.; Rinn, J.L.; Pachter, L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat. Biotechnol. 2013, 31, 46–53. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |




| Item | No. of Sequences |
|---|---|
| High-quality reads | 929,225,004 |
| Average length (bp) | 9,208,092,085 |
| No. of contig > 500 bp | 91,092 |
| Total unigenes | 182,881 |
| N50 contig size (bp) | 1374 |
| Total assemble bases (bp) | 156,275,266 |
© 2017 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zhang, J.-J.; Su, H.; Zhang, L.; Liao, B.-S.; Xiao, S.-M.; Dong, L.-L.; Hu, Z.-G.; Wang, P.; Li, X.-W.; Huang, Z.-H.; et al. Comprehensive Characterization for Ginsenosides Biosynthesis in Ginseng Root by Integration Analysis of Chemical and Transcriptome. Molecules 2017, 22, 889. https://doi.org/10.3390/molecules22060889
Zhang J-J, Su H, Zhang L, Liao B-S, Xiao S-M, Dong L-L, Hu Z-G, Wang P, Li X-W, Huang Z-H, et al. Comprehensive Characterization for Ginsenosides Biosynthesis in Ginseng Root by Integration Analysis of Chemical and Transcriptome. Molecules. 2017; 22(6):889. https://doi.org/10.3390/molecules22060889
Chicago/Turabian StyleZhang, Jing-Jing, He Su, Lei Zhang, Bao-Sheng Liao, Shui-Ming Xiao, Lin-Lin Dong, Zhi-Gang Hu, Ping Wang, Xi-Wen Li, Zhi-Hai Huang, and et al. 2017. "Comprehensive Characterization for Ginsenosides Biosynthesis in Ginseng Root by Integration Analysis of Chemical and Transcriptome" Molecules 22, no. 6: 889. https://doi.org/10.3390/molecules22060889
APA StyleZhang, J. -J., Su, H., Zhang, L., Liao, B. -S., Xiao, S. -M., Dong, L. -L., Hu, Z. -G., Wang, P., Li, X. -W., Huang, Z. -H., Gao, Z. -M., Zhang, L. -J., Shen, L., Cheng, R. -Y., Xu, J., & Chen, S. -L. (2017). Comprehensive Characterization for Ginsenosides Biosynthesis in Ginseng Root by Integration Analysis of Chemical and Transcriptome. Molecules, 22(6), 889. https://doi.org/10.3390/molecules22060889