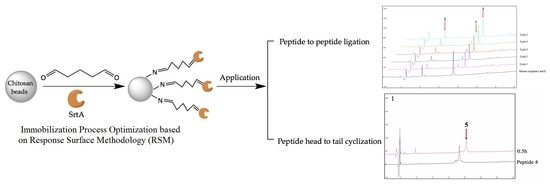
Immobilization of Staphylococcus aureus Sortase A on Chitosan Particles and Its Applications in Peptide-to-Peptide Ligation and Peptide Cyclization
Abstract
:1. Introduction
2. Results and Discussion
2.1. Characterization of SrtA Immobilized Chitosan Particles
2.2. Process Optimization of Immobilization of SrtA onto Chitosan Particles
2.2.1. Identification of the Significant Variables Using the Plackett–Burman (PB) Design
2.2.2. Optimization of Operational Parameters by the Box–Behnken Experimental Design (BBD)
2.3. pH and Thermo-Stability of Immobilized SrtA
2.4. Immobilized SrtA-Mediated Peptide-to-Peptide Ligation
2.5. Immobilized SrtA-Mediated Peptide Cyclization and Its Scale-Up Capability
3. Experimental Section
3.1. Materials and Methods
3.1.1. Materials
3.1.2. Expression and Purification of SrtA
3.1.3. Preparation of Chitosan Particles
3.1.4. Immobilization of SrtA
3.1.5. Chitosan Particles Characterization
3.2. Experimental Design and Data Analysis
3.2.1. Plackett–Burman Experimental Design
3.2.2. Response Surface Methodology (RSM)
3.3. Activity Assays of SrtA
3.4. Determination of Protein Concentration
3.5. Stability of Free and Immobilized SrtA
3.5.1. pH Stability
3.5.2. Thermal Stability
3.6. Application of Immobilized SrtA in Peptide-to-Peptide Ligation and Peptide Cyclization
3.6.1. Immobilized SrtA-Mediated Peptide-to-Peptide Ligation
3.6.2. Immobilized SrtA-Mediated Peptide Cyclization
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Mazmanian, S.K.; Liu, G.; Ton-That, H.; Schneewind, O. Staphylococcus aureus sortase, an enzyme that anchors surface proteins to the cell wall. Science 1999, 285, 760–763. [Google Scholar] [CrossRef] [PubMed]
- Ton-That, H.; Liu, G.; Mazmanian, S.K.; Faull, K.F.; Schneewind, O. Purification and characterization of sortase, the transpeptidase that cleaves surface proteins of Staphylococcus aureus at the LPXTG motif. Proc. Natl. Acad. Sci. USA 1999, 96, 12424–12429. [Google Scholar] [CrossRef] [PubMed]
- Ton-That, H.; Mazmanian, S.K.; Faull, K.F.; Schneewind, O. Anchoring of surface proteins to the cell wall of Staphylococcus aureus. Sortase catalyzed in vitro transpeptidation reaction using LPXTG peptide and NH2-Gly3 substrates. J. Biol. Chem. 2000, 275, 9876–9881. [Google Scholar] [CrossRef] [PubMed]
- Mao, H.; Hart, S.A.; Schink, A.; Pollok, B.A. Sortase-mediated protein ligation: A new method for protein engineering. J. Am. Chem. Soc. 2004, 126, 2670–2671. [Google Scholar] [CrossRef] [PubMed]
- Parthasarathy, R.; Subramanian, S.; Boder, E.T. Sortase A as a novel molecular “stapler” for sequence-specific protein conjugation. Bioconjug. Chem. 2007, 18, 469–476. [Google Scholar] [CrossRef] [PubMed]
- Levary, D.A.; Parthasarathy, R.; Boder, E.T.; Ackerman, M.E. Protein-protein fusion catalyzed by sortase A. PLoS ONE 2011, 6, e18342. [Google Scholar] [CrossRef] [PubMed]
- Wagner, K.; Kwakkenbos, M.J.; Claassen, Y.B.; Maijoor, K.; Bohne, M.; van der Sluijs, K.F.; Witte, M.D.; van Zoelen, D.J.; Cornelissen, L.A.; Beaumont, T.; et al. Bispecific antibody generated with sortase and click chemistry has broad antiinfluenza virus activity. Proc. Natl. Acad. Sci. USA 2014, 111, 16820–16825. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Zhang, R.; Xu, Y.; Wu, Z.; Li, J.; Zhou, X.; Jiang, J.; Liu, H.; Xiao, R. Sortase A-mediated crosslinked short-chain dehydrogenases/reductases as novel biocatalysts with improved thermostability and catalytic efficiency. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Antos, J.M.; Miller, G.M.; Grotenbreg, G.M.; Ploegh, H.L. Lipid modification of proteins through sortase-catalyzed transpeptidation. J. Am. Chem. Soc. 2008, 130, 16338–16343. [Google Scholar] [CrossRef] [PubMed]
- Glasgow, J.E.; Salit, M.L.; Cochran, J.R. In Vivo Site-Specific Protein Tagging with Diverse Amines Using an Engineered Sortase Variant. J. Am. Chem. Soc. 2016, 138, 7496–7499. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Guo, X.; Guo, Z. Sortase A-catalyzed peptide cyclization for the synthesis of macrocyclic peptides and glycopeptides. Chem. Commun. 2011, 47, 9218–9220. [Google Scholar] [CrossRef] [PubMed]
- Bolscher, J.G.; Oudhoff, M.J.; Nazmi, K.; Antos, J.M.; Guimaraes, C.P.; Spooner, E.; Haney, E.F.; Garcia Vallejo, J.J.; Vogel, H.J.; van’t Hof, W.; et al. Sortase A as a tool for high-yield histatin cyclization. FASEB J. 2011, 25, 2650–2658. [Google Scholar] [CrossRef] [PubMed]
- Jia, X.; Kwon, S.; Wang, C.I.; Huang, Y.H.; Chan, L.Y.; Tan, C.C.; Rosengren, K.J.; Mulvenna, J.P.; Schroeder, C.I.; Craik, D.J. Semienzymatic cyclization of disulfide-rich peptides using sortase A. J. Biol. Chem. 2014, 289, 6627–6638. [Google Scholar] [CrossRef] [PubMed]
- Park, K.; Jung, J.; Son, J.; Kim, S.H.; Chung, B.H. Anchoring foreign substances on live cell surfaces using sortase a specific binding peptide. Chem. Commun. 2013, 49, 9585–9587. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, T.; Yamamoto, T.; Tsukiji, S.; Nagamune, T. Site-specific protein modification on living cells catalyzed by sortase. Chembiochem 2008, 9, 802–807. [Google Scholar] [CrossRef] [PubMed]
- Guo, X.; Wu, Z.; Guo, Z. New method for site-specific modification of liposomes with proteins using sortase A-mediated transpeptidation. Bioconj. Chem. 2012, 23, 650–655. [Google Scholar] [CrossRef] [PubMed]
- Jiang, R.; Weingart, J.; Zhang, H.; Ma, Y.; Sun, X.L. End-point immobilization of recombinant thrombomodulin via sortase-mediated ligation. Bioconj. Chem. 2012, 23, 643–649. [Google Scholar] [CrossRef] [PubMed]
- Ham, H.O.; Qu, Z.; Haller, C.A.; Dorr, B.M.; Dai, E.; Kim, W.; Liu, D.R.; Chaikof, E.L. In situ regeneration of bioactive coatings enabled by an evolved Staphylococcus aureus sortase A. Nat. Commun. 2016, 7. [Google Scholar] [CrossRef] [PubMed]
- Schmohl, L.; Schwarzer, D. Sortase-mediated ligations for the site-specific modification of proteins. Curr. Opin. Chem. Biol. 2014, 22, 122–128. [Google Scholar] [CrossRef] [PubMed]
- Antos, J.M.; Truttmann, M.C.; Ploegh, H.L. Recent advances in sortase-catalyzed ligation methodology. Curr. Opin. Struct. Biol. 2016, 38, 111–118. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Guo, Z. Sortase-mediated transpeptidation for site-specific modification of peptides, glycopeptides, and proteins. J. Carbohydr. Chem. 2012, 31, 48–66. [Google Scholar] [CrossRef] [PubMed]
- Tsukiji, S.; Nagamune, T. Sortase-mediated ligation: A gift from gram-positive bacteria to protein engineering. Chembiochem 2009, 10, 787–798. [Google Scholar] [CrossRef] [PubMed]
- Beerli, R.R.; Hell, T.; Merkel, A.S.; Grawunder, U. Sortase enzyme-mediated generation of site-specifically conjugated antibody drug conjugates with high in vitro and in vivo potency. PLoS ONE 2015, 10, e0131177. [Google Scholar] [CrossRef] [PubMed]
- Pan, L.; Zhao, W.; Lai, J.; Ding, D.; Zhang, Q.; Yang, X.; Huang, M.; Jin, S.; Xu, Y.; Zeng, S.; et al. Sortase A-generated highly potent anti-CD20-MMAE conjugates for efficient elimination of B-lineage lymphomas. Small 2017, 13. [Google Scholar] [CrossRef] [PubMed]
- Kruger, R.G.; Otvos, B.; Frankel, B.A.; Bentley, M.; Dostal, P.; McCafferty, D.G. Analysis of the substrate specificity of the Staphylococcus aureus sortase transpeptidase SrtA. Biochemistry 2004, 43, 1541–1551. [Google Scholar] [CrossRef] [PubMed]
- Chen, I.; Dorr, B.M.; Liu, D.R. A general strategy for the evolution of bond-forming enzymes using yeast display. Proc. Natl. Acad. Sci. USA 2011, 108, 11399–11404. [Google Scholar] [CrossRef] [PubMed]
- Chen, L.; Cohen, J.; Song, X.; Zhao, A.; Ye, Z.; Feulner, C.J.; Doonan, P.; Somers, W.; Lin, L.; Chen, P.R. Improved variants of SrtA for site-specific conjugation on antibodies and proteins with high efficiency. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.; Hong, H.; Zhao, X.; Wang, X. Efficient expression of sortase a from Staphylococcus aureus in Escherichia coli and its enzymatic characterizations. Bioresour. Bioprocess. 2017, 4. [Google Scholar] [CrossRef] [PubMed]
- DiCosimo, R.; McAuliffe, J.; Poulose, A.J.; Bohlmann, G. Industrial use of immobilized enzymes. Chem. Soc. Rev. 2013, 42, 6437–6474. [Google Scholar] [CrossRef] [PubMed]
- Sheldon, R.A.; van Pelt, S. Enzyme immobilisation in biocatalysis: Why, what and how. Chem. Soc. Rev. 2013, 42, 6223–6235. [Google Scholar] [CrossRef] [PubMed]
- Rodrigues, R.C.; Hernandez, K.; Barbosa, O.; Rueda, N.; Garcia-Galan, C.; dos Santos, J.C.S.; Berenguer-Murcia, A.; Fernandez-Lafuente, R. Immobilization of proteins in poly-styrene-divinylbenzene matrices: Functional properties and applications. Curr. Org. Chem. 2015, 19, 1707–1718. [Google Scholar] [CrossRef]
- Steinhagen, M.; Zunker, K.; Nordsieck, K.; Beck-Sickinger, A.G. Large scale modification of biomolecules using immobilized sortase A from Staphylococcus aureus. Bioorg. Med. Chem. 2013, 21, 3504–3510. [Google Scholar] [CrossRef] [PubMed]
- Policarpo, R.L.; Kang, H.; Liao, X.; Rabideau, A.E.; Simon, M.D.; Pentelute, B.L. Flow-based enzymatic ligation by sortase A. Angew. Chem. 2014, 53, 9203–9208. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Hong, H.; Cheng, X.; Liu, S.; Deng, T.; Guo, Z.; Wu, Z. One-step purification and immobilization of extracellularly expressed sortase A by magnetic particles to develop a robust and recyclable biocatalyst. Sci. Rep. 2017, 7. [Google Scholar] [CrossRef] [PubMed]
- Witte, M.D.; Wu, T.; Guimaraes, C.P.; Theile, C.S.; Blom, A.E.; Ingram, J.R.; Li, Z.; Kundrat, L.; Goldberg, S.D.; Ploegh, H.L. Site-specific protein modification using immobilized sortase in batch and continuous-flow systems. Nat. Protoc. 2015, 10, 508–516. [Google Scholar] [CrossRef] [PubMed]
- Rosen, C.B.; Kwant, R.L.; MacDonald, J.I.; Rao, M.; Francis, M.B. Capture and recycling of sortase A through site-specific labeling with Lithocholic acid. Angew. Chem. 2016, 55, 8585–8589. [Google Scholar] [CrossRef] [PubMed]
- Biro, E.; Nemeth, A.S.; Sisak, C.; Feczko, T.; Gyenis, J. Preparation of chitosan particles suitable for enzyme immobilization. J. Biochem. Biophys. Methods 2008, 70, 1240–1246. [Google Scholar] [CrossRef] [PubMed]
- Krajewska, B. Application of chitin- and chitosan-based materials for enzyme immobilizations: A review. Enzym. Microb. Technol. 2004, 35, 126–139. [Google Scholar] [CrossRef]
- Vassiliadi, E.; Xenakis, A.; Zoumpanioti, M. Chitosan hydrogels: A new and simple matrix for lipase catalysed biosyntheses. Mol. Catal. 2018, 445, 206–212. [Google Scholar] [CrossRef]
- Dos Santos, J.C.S.; Bonazza, H.L.; de Matos, L.; Carneiro, E.A.; Barbosa, O.; Fernandez-Lafuente, R.; Goncalves, L.R.B.; de Sant’ Ana, H.B.; Santiago-Aguiar, R.S. Immobilization of CALB on activated chitosan: Application to enzymatic synthesis in supercritical and near-critical carbon dioxide. Biotechnol. Rep. 2017, 14, 16–26. [Google Scholar] [CrossRef] [PubMed]
- Klein, M.P.; Hackenhaar, C.R.; Lorenzoni, A.S.G.; Rodrigues, R.C.; Costa, T.M.H.; Ninow, J.L.; Hertz, P.F. Chitosan crosslinked with genipin as support matrix for application in food process: Support characterization and beta-d-galactosidase immobilization. Carbohydr. Polym. 2016, 137, 184–190. [Google Scholar] [CrossRef] [PubMed]
- Cappannella, E.; Benucci, I.; Lombardelli, C.; Liburdi, K.; Bavaro, T.; Esti, M. Immobilized lysozyme for the continuous lysis of lactic bacteria in wine: Bench-scale fluidized-bed reactor study. Food Chem. 2016, 210, 49–55. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Zhu, K.X.; Zhou, H.M. Immobilization of glucose oxidase in alginate-chitosan microcapsules. Int. J. Mol. Sci. 2011, 12, 3042–3054. [Google Scholar] [CrossRef] [PubMed]
- Skoronski, E.; Fernandes, M.; Magalhaes, M.D.B.; da Silva, G.F.; Joao, J.J.; Soares, C.H.L.; Furigo, A. Substrate Specificity and Enzyme Recycling Using Chitosan Immobilized Laccase. Molecules 2014, 19, 16794–16809. [Google Scholar] [CrossRef] [PubMed]
- Barbosa, O.; Ortiz, C.; Berenguer-Murcia, A.; Torres, R.; Rodrigues, R.C.; Fernandez-Lafuente, R. Glutaraldehyde in bio-catalysts design: A useful crosslinker and a versatile tool in enzyme immobilization. RSC Adv. 2014, 4, 1583–1600. [Google Scholar] [CrossRef]
- Fernandez-Lafuente, R.; Rosell, C.M.; Rodriguez, V.; Guisan, J.M. Strategies for enzyme stabilization by intramolecular crosslinking with bifunctional reagents. Enzym. Microb. Technol. 1995, 17, 517–523. [Google Scholar] [CrossRef]
- Banerjee, A.; Banerjee, S.; Sarkar, P. Statistical design of experiments for optimization of arsenate reductase production by Kocuria palustris (RJB-6) and immobilization parameters in polymer beads. RSC Adv. 2016, 6, 49289–49297. [Google Scholar] [CrossRef]
- Srivastava, P.K.; Anand, A. Immobilization of acid phosphatase from Vigna aconitifolia seeds on chitosan beads and its characterization. Int. J. Biol. Macromol. 2014, 64, 150–154. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.N.; Singh, S.; Suthar, N.; Dubey, V.K. Glutaraldehyde-activated chitosan matrix for immobilization of a novel cysteine protease, Procerain B. J. Agric. Food Chem. 2011, 59, 6256–6262. [Google Scholar] [CrossRef] [PubMed]
- De Oliveira, U.M.F.; Lima de Matos, L.J.B.; de Souza, M.C.M.; Pinheiro, B.B.; Dos Santos, J.C.S.; Goncalves, L.R.B. Effect of the presence of surfactants and immobilization conditions on catalysts’ properties of Rhizomucor miehei lipase onto chitosan. Appl. Biochem. Biotechnol. 2017. [Google Scholar] [CrossRef] [PubMed]
- Urrutia, P.; Arrieta, R.; Alvarez, L.; Cardenas, C.; Mesa, M.; Wilson, L. Immobilization of lipases in hydrophobic chitosan for selective hydrolysis of fish oil: The impact of support functionalization on lipase activity, selectivity and stability. Int. J. Biol. Macromol. 2017, 108, 674–686. [Google Scholar] [CrossRef] [PubMed]
- Kwon, S.; Bosmans, F.; Kaas, Q.; Cheneval, O.; Conibear, A.C.; Rosengren, K.J.; Wang, C.K.; Schroeder, C.I.; Craik, D.J. Efficient enzymatic cyclization of an inhibitory cystine knot-containing peptide. Biotechnol. Bioeng. 2016, 113, 2202–2212. [Google Scholar] [CrossRef] [PubMed]
- Van’t Hof, W.; Hansenova Manaskova, S.; Veerman, E.C.; Bolscher, J.G. Sortase-mediated backbone cyclization of proteins and peptides. Biol. Chem. 2015, 396, 283–293. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.M.; Liu, S.Z.; Cheng, X.Z.; Zhao, X.R.; Hong, H.F. High yield synthesis of cyclic analogues of antibacterial peptides P-113 by Sortase A-mediated ligation and their conformation studies. Chin. Chem. Lett. 2017, 28, 553–557. [Google Scholar] [CrossRef]
- Cavello, I.A.; Contreras-Esquivel, J.C.; Cavalitto, S.F. Immobilization of a keratinolytic protease from Purpureocillium lilacinum on genipin activated-chitosan particles. Process Biochem. 2014, 49, 1332–1336. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |







| Effect Value | F Value | Prob > F | Value | |
|---|---|---|---|---|
| A | −16.29 | 0.18 | 0.6949 | - |
| B | −345.78 | 80.14 | 0.0009 | - |
| C | −75.89 | 3.86 | 0.1209 | - |
| D | 92.75 | 5.77 | 0.0743 | - |
| E | −211.76 | 30.06 | 0.0054 | - |
| F | 110.39 | 8.17 | 0.0460 | - |
| G | 164.69 | 18.18 | 0.0130 | - |
| Model | - | 20.91 | 0.0053 | - |
| R2 | - | - | - | 0.97 |
| Adj R2 | - | - | - | 0.93 |
| Pred R2 | - | - | - | 0.76 |
| A (%) | B (mg·mL−1) | C (h) | |
|---|---|---|---|
| Low level (−1) | 0.2 | 0.2 | 4 |
| Central level (0) | 0.5 | 0.5 | 8 |
| High level (+1) | 0.8 | 0.8 | 12 |
| Entry | A (%) | B (mg·mL−1) | C (h) | Total Enzyme Activity | Binding Protein (mg) | Loading Efficiency (%) | Specific Activity (U·mg−1) | Activity Retention (AR) (%) |
|---|---|---|---|---|---|---|---|---|
| 1 | −1 | 0 | +1 | 97.55 | 0.098 | 39.2 | 998.65 | 3.21 |
| 2 | 0 | 0 | 0 | 588.28 | 0.199 | 79.6 | 2952.47 | 9.50 |
| 3 | −1 | 0 | −1 | 79.05 | 0.067 | 26.8 | 1172.03 | 3.77 |
| 4 | −1 | +1 | 0 | 167.93 | 0.151 | 37.8 | 1115.51 | 3.59 |
| 5 | 0 | −1 | +1 | 165.85 | 0.100 | 100 | 1658.50 | 5.33 |
| 6 | +1 | 0 | +1 | 346.48 | 0.250 | 100 | 1385.93 | 4.46 |
| 7 | 0 | 0 | 0 | 550.10 | 0.185 | 74 | 2971.93 | 9.56 |
| 8 | 0 | +1 | +1 | 849.78 | 0.309 | 77.3 | 2751.93 | 8.85 |
| 9 | 0 | −1 | −1 | 276.50 | 0.085 | 85 | 3242.40 | 10.43 |
| 10 | 0 | 0 | 0 | 637.48 | 0.185 | 74 | 3438.04 | 11.06 |
| 11 | −1 | −1 | 0 | 53.75 | 0.037 | 37 | 1451.57 | 4.67 |
| 12 | 0 | +1 | −1 | 665.40 | 0.255 | 63.8 | 2611.89 | 8.40 |
| 13 | +1 | 0 | −1 | 450.12 | 0.221 | 88.4 | 2035.59 | 6.55 |
| 14 | +1 | +1 | 0 | 599.80 | 0.356 | 89 | 1686.33 | 5.42 |
| 15 | +1 | −1 | 0 | 170.65 | 0.099 | 99 | 1721.87 | 5.54 |
| 16 | 0 | 0 | 0 | 592.40 | 0.182 | 72.8 | 3251.30 | 10.46 |
| 17 | 0 | 0 | 0 | 572.47 | 0.185 | 74 | 3098.15 | 9.96 |
| F Value | Prob > F | Value | ||
|---|---|---|---|---|
| Model | 27.75 | 0.0001 | - | Significant |
| A | 11.84 | 0.0108 | - | - |
| B | 0.023 | 0.8848 | - | - |
| C | 13.90 | 0.0074 | - | - |
| AB | 0.49 | 0.5071 | - | - |
| AC | 1.23 | 0.3045 | - | - |
| BC | 16.08 | 0.0051 | - | - |
| A2 | 180.52 | <0.0001 | - | - |
| B2 | 5.22 | 0.0563 | - | - |
| C2 | 10.23 | 0.0151 | - | - |
| R2 | - | - | 0.9727 | - |
| Adj R2 | - | - | 0.9377 | - |
| Pred R2 | - | - | 0.7673 | - |
| Lack of fit | 1.25 | 0.4040 | Not significant |
| Entry | Volume (0.48 mg/mL−1) | Reaction Time (h) | Number of Beads | Yield (%) |
|---|---|---|---|---|
| 1 | 2 mL | 0.5 | 40 | 100 |
| 2 | 10 mL | 0.5 | 200 | 98.5 |
| 3 | 20 mL | 1 | 200 | 97.3 |
| 4 | 40 mL | 2 | 400 | 95.4 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, M.; Hong, H.; Liu, S.; Zhao, X.; Wu, Z. Immobilization of Staphylococcus aureus Sortase A on Chitosan Particles and Its Applications in Peptide-to-Peptide Ligation and Peptide Cyclization. Molecules 2018, 23, 192. https://doi.org/10.3390/molecules23010192
Yang M, Hong H, Liu S, Zhao X, Wu Z. Immobilization of Staphylococcus aureus Sortase A on Chitosan Particles and Its Applications in Peptide-to-Peptide Ligation and Peptide Cyclization. Molecules. 2018; 23(1):192. https://doi.org/10.3390/molecules23010192
Chicago/Turabian StyleYang, Min, Haofei Hong, Shaozhong Liu, Xinrui Zhao, and Zhimeng Wu. 2018. "Immobilization of Staphylococcus aureus Sortase A on Chitosan Particles and Its Applications in Peptide-to-Peptide Ligation and Peptide Cyclization" Molecules 23, no. 1: 192. https://doi.org/10.3390/molecules23010192
APA StyleYang, M., Hong, H., Liu, S., Zhao, X., & Wu, Z. (2018). Immobilization of Staphylococcus aureus Sortase A on Chitosan Particles and Its Applications in Peptide-to-Peptide Ligation and Peptide Cyclization. Molecules, 23(1), 192. https://doi.org/10.3390/molecules23010192