Probing the Structural Determinants of Amino Acid Recognition: X-Ray Studies of Crystalline Ditopic Host-Guest Complexes of the Positively Charged Amino Acids, Arg, Lys, and His with a Cavitand Molecule
Abstract
:1. Introduction
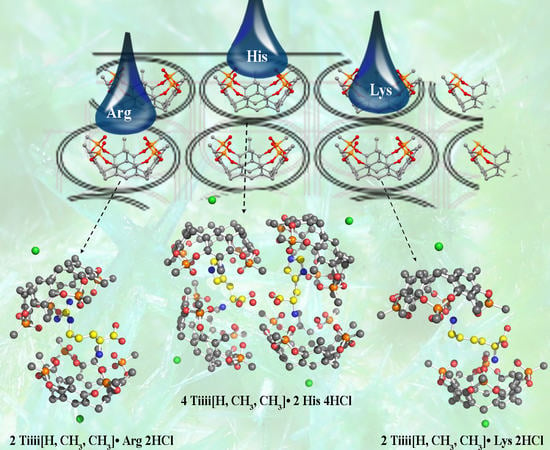
2. Results and Discussion
3. Materials and Methods
3.1. Co-Crystallization Experiments
3.2. Crystal Structure Determination
3.2.1. Structure Refinement of 2 Tiiii[H, CH3, CH3]•Arg 2HCl Complex
3.2.2. Structure Refinement of 2 Tiiii[H, CH3, CH3]•Lys 2HCl Complex
3.2.3. Structure Refinement of 2 Tiiii[H, CH3, CH3]•His 2HCl Complex
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Tapiero, H.; Mathé, G.; Couvreur, P.; Tew, K.D. Arginine. Biomed. Pharmacother. 2002, 56, 439–445. [Google Scholar] [CrossRef]
- Stechmiller, J.K.; Childress, B.; Cowan, L. Arginine Supplementation and Wound Healing. Nutr. Clin. Pract. 2005, 20, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Vila, J.A.; Ripoll, D.R.; Villegas, M.E.; Vorobjev, Y.N.; Scheraga, H.A. Role of Hydrophobicity and Solvent-Mediated Charge-Charge Interactions in Stabilizing α-Helices. BioPhys. J. 1998, 75, 2637–2646. [Google Scholar] [CrossRef]
- Strickler, S.S.; Gribenko, A.V.; Gribenko, A.V.; Keiffer, T.R.; Tomlinson, J.; Reihle, T.; Loladze, V.V.; Makhatadze, G.I. Protein Stability and Surface Electrostatics: A Charged Relationship. Biochemistry 2006, 45, 2761–2766. [Google Scholar] [CrossRef]
- Borders, C.L., Jr.; Broadwater, J.A.; Bekeny, P.A.; Salmon, J.E.; Lee, A.S.; Eldridge, A.M.; Pett, V.B. A structural role for arginine in proteins: Multiple hydrogen bonds to backbone carbonyl oxygens. Protein Sci. 1994, 3, 541–548. [Google Scholar] [CrossRef]
- Tsukada, Y.; Fang, J.; Erdjument-Bromage, H.; Warren, M.E.; Borchers, C.H.; Tempst, P.; Zhang, Y. Histone demethylation by a family of JmjC domain-containing proteins. Nature 2006, 439, 811–816. [Google Scholar] [CrossRef]
- Stewart, M.D.; Li, J.; Wong, J. Relationship between Histone H3 Lysine 9 Methylation, Transcription Repression, and Heterochromatin Protein 1 Recruitment. J. Mol. Cell. Biol. 2005, 25, 2525–2538. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakayama, J.; Rice, J.C.; Strahl, B.D.; Allis, C.D.; Grewal, S.I. Role of Histone H3 Lysine 9 Methylation in Epigenetic Control of Heterochromatin Assembly. Science 2001, 292, 110–113. [Google Scholar] [CrossRef] [PubMed]
- Chen, G.N.; Wu, X.P.; Duan, J.P.; Chen, H.Q. A study on electrochemistry of histidine and its metabolites based on the diazo coupling reaction. Talanta 1999, 49, 319–330. [Google Scholar]
- Liao, S.-M.; Du, Q.-S.; Meng, J.-Z.; Pang, Z.-W.; Huang, R.-B. The multiple roles of histidine in protein interactions. Chem. Cent. J. 2013, 7, 44. [Google Scholar] [CrossRef] [Green Version]
- Beadle, C.; Long, G.W.; Weiss, W.R.; McElroy, P.D.; Maret, S.M.; Oloo, A.J.; Hoffman, S.L. Diagnosis of malaria by detection of Plasmodium falciparum HRP-2 antigen with a rapid dipstick antigen-capture assay. Lancet 1994, 343, 564–568. [Google Scholar] [CrossRef]
- Chen, H.; Gu, L.; Yin, Y.; Koh, K.; Lee, J. Molecular Recognition of Arginine by Supramolecular Complexation with Calixarene Crown Ether Based on Surface Plasmon Resonance. Int. J. Mol. Sci. 2011, 12, 2315–2324. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fokkens, M.; Schrader, T.; Klärner, F.-G. A Molecular Tweezer for Lysine and Arginine. J. Am. Chem. Soc. 2005, 127, 14415–14421. [Google Scholar] [CrossRef] [PubMed]
- Ngola, S.M.; Kearney, P.C.; Mecozzi, S.; Russell, K.; Dougherty, D.A. A Selective Receptor for Arginine Derivatives in Aqueous Media. Energetic Consequences of Salt Bridges that Are Highly Exposed to Water. J. Am. Chem. Soc. 1999, 121, 1192–1201. [Google Scholar] [CrossRef]
- Julian, R.R.; Akin, M.; May, J.A.; Stoltz, B.M.; Beauchamp, J.L. Molecular recognition of arginine in small peptides by supramolecular complexation with dibenzo-30-crown-10 ether. Int. J. Mass Spectrom. 2002, 220, 87–96. [Google Scholar] [CrossRef]
- Chen, Z.; Liu, J.; Han, Y.; Zhu, L. A novel histidine assay using tetraphenylporphyrin manganese (III) chloride as a molecular recognition probe by resonance light scattering technique. Anal. Chim. Acta 2006, 570, 109–115. [Google Scholar] [CrossRef]
- Cram, D.J.; Cram, J.M. Container Molecules and Their Guests; Stoddart, J.F., Ed.; The Royal Society of Chemistry: Cambridge, UK, 1994. [Google Scholar]
- Pinalli, R.; Pedrini, A.; Dalcanale, E. Cavitands. In Comprehensive Supramolecular Chemistry II; Atwood, J.L., Ed.; Elsevier: Amsterdam, The Netherlands, 2017; pp. 87–115. ISBN 9780128031995. [Google Scholar]
- Pinalli, R.; Dalcanale, E.; Ugozzoli, F.; Massera, C. Resorcinarene-based cavitands as building blocks for crystal engineering. CrystEngComm 2016, 18, 5788–5802. [Google Scholar] [CrossRef]
- Rebek, J., Jr. Molecular Recognition with Model Systems. Angew. Chem. Int. Ed. Engl. 1990, 29, 245–255. [Google Scholar] [CrossRef]
- Hunter, C.H.; Lawson, K.R.; Perkins, J.; Urch, C.J. Aromatic interactions. J. Chem. Soc., Perkin Trans. 2001, 2, 651–669. [Google Scholar] [CrossRef]
- Nishio, M.; Hirota, M.; Umezawa, Y. The CH-π Interactions: Evidence, Nature and Consequences; Wiley-VCH: New York, NY, USA, 1998. [Google Scholar]
- Dougherty, D.A. The Cation-π Interaction. Acc. Chem. Res. 2013, 46, 885–893. [Google Scholar] [CrossRef]
- Bianchi, F.; Mattarozzi, M.; Betti, P.; Bisceglie, F.; Careri, M.; Mangia, A.; Sidisky, L.; Ongarato, S.; Dalcanale, E. Innovative Cavitand-Based Sol-Gel Coatings for the Environmental Monitoring of Benzene and Chlorobenzenes via Solid-Phase Microextraction. Anal. Chem. 2008, 80, 6423–6430. [Google Scholar] [CrossRef]
- Tudisco, C.; Fragalà, M.E.; Giuffrida, A.E.; Bertani, F.; Pinalli, R.; Dalcanale, E.; Compagnini, G.; Condorelli, G.G. Hierarchical Route for the Fabrication of Cavitand-Modified Nanostructured ZnO Fibers for Volatile Organic Compound Detection. J. Phys. Chem. C 2016, 120, 12611–12617. [Google Scholar] [CrossRef]
- Trzciński, J.W.; Pinalli, R.; Riboni, N.; Pedrini, A.; Bianchi, F.; Zampolli, S.; Elmi, I.; Massera, C.; Ugozzoli, F.; Dalcanale, E. In Search of the Ultimate Benzene Sensor: The EtQxBox Solution. ACS Sensors 2017, 2, 590–598. [Google Scholar] [CrossRef] [PubMed]
- Pinalli, R.; Pedrini, A.; Dalcanale, E. Environmental gas sensing with cavitands. Chem Eur. J. 2018, 24, 1010–1019. [Google Scholar] [CrossRef]
- Giannetto, M.; Pedrini, A.; Fortunati, S.; Brando, D.; Milano, S.; Massera, C.; Tatti, R.; Verucchi, R.; Careri, M.; Dalcanale, E.; et al. Sensing of halogenated aromatic hydrocarbons in water with a cavitand coated piezoelectric device. Sens. Actuators B Chem. 2018, 276, 340–348. [Google Scholar] [CrossRef]
- Maffei, F.; Betti, P.; Genovese, D.; Montalti, M.; Prodi, L.; De Zorzi, R.; Geremia, S.; Dalcanale, E. Highly Selective Chemical Vapor Sensing by Molecular Recognition: Specific Detection of C1–C4 Alcohols with a Fluorescent Phosphonate Cavitand. Angew. Chem. Int. Ed. 2011, 50, 4654–4657. [Google Scholar] [CrossRef]
- Melegari, M.; Massera, C.; Pinalli, R.; Yebeutchou, R.M.; Dalcanale, E. Supramolecular sensing of short chain alcohols with mixed-bridged thio-phosphonate cavitands. Sens. Actuators B Chem. 2013, 179, 74–80. [Google Scholar] [CrossRef]
- Dionisio, M.; Oliviero, G.; Menozzi, D.; Federici, S.; Yebeutchou, R.M.; Schmidtchen, F.P.; Dalcanale, E.; Bergese, P. Nanomechanical Recognition of N-Methylammonium Salts. J. Am. Chem. Soc. 2012, 134, 2392–2398. [Google Scholar] [CrossRef]
- Pinalli, R.; Suman, M.; Dalcanale, E. Cavitands at work: From molecular recognition to supramolecular sensors. Eur. J. Org. Chem. 2004, 3, 451–462. [Google Scholar] [CrossRef]
- Biavardi, E.; Tudisco, C.; Maffei, F.; Motta, A.; Massera, C.; Condorelli, G.G.; Dalcanale, E. Exclusive recognition of sarcosine in water and urine by a cavitand-functionalized silicon surface. Proc. Natl. Acad. Sci. USA 2012, 109, 2263–2268. [Google Scholar] [CrossRef] [Green Version]
- Valenti, G.; Rampazzo, E.; Biavardi, E.; Villani, E.; Fracasso, G.; Marcaccio, M.; Bertani, F.; Ramarli, D.; Dalcanale, E.; Paolucci, F.; et al. An electrochemiluminescence-supramolecular approach to sarcosine detection for early diagnosis of prostate cancer. Faraday Discuss. 2015, 185, 299–309. [Google Scholar] [CrossRef] [PubMed]
- Pinalli, R.; Brancatelli, G.; Pedrini, A.; Menozzi, D.; Hernández, D.; Ballester, P.; Geremia, S.; Dalcanale, E. The origin of selectivity in the complexation of N-methyl amino acids by tetraphosphonate cavitands. J. Am. Chem. Soc. 2016, 138, 8569–8580. [Google Scholar] [CrossRef]
- Liu, Y.; Perez, L.; Mettry, M.; Easley, C.J.; Hooley, R.J.; Zhong, W. Self-Aggregating Deep Cavitand Acts as a Fluorescence Displacement Sensor for Lysine Methylation. J. Am. Chem. Soc. 2016, 138, 10746–10749. [Google Scholar] [CrossRef]
- Liu, Y.; Perez, L.; Gill, A.D.; Mettry, M.; Li, L.; Wang, Y.; Hooley, R.J.; Zhong, W. Site-Selective Sensing of Histone Methylation Enzyme Activity via an Arrayed Supramolecular Tandem Assay. J. Am. Chem. Soc. 2017, 139, 10964–10967. [Google Scholar] [CrossRef]
- Pinalli, R.; Pedrini, A.; Dalcanale, E. Biochemical Sensing with cavitands. Chem. Soc. Rev. 2018, 47, 7006–7026. [Google Scholar] [CrossRef] [PubMed]
- McGovern, R.E.; Fernandes, H.; Khan, A.R.; Power, N.P.; Crowley, P.B. Protein camouflage in cytochrome c-calixarene complexes. Nat. Chem. 2012, 4, 527–533. [Google Scholar] [CrossRef]
- Rennie, M.L.; Fox, G.C.; Pérez, J.; Crowley, P.B. Auto-regulated Protein Assembly on a Supramolecular Scaffold. Angew. Chem. Int. Ed. 2018, 57, 13764–13769. [Google Scholar] [CrossRef]
- McGovern, R.E.; McCarthy, A.A.; Crowley, P.B. Protein assembly mediated by sulfonatocalix[4]arene. Chem. Commun. 2014, 50, 10412–10415. [Google Scholar] [CrossRef]
- Chinai, J.M.; Taylor, A.B.; Ryno, L.M.; Hargreaves, N.D.; Morris, C.A.; Hart, P.J.; Urbach, A.R. Molecular recognition of insulin by a synthetic receptor. J. Am. Chem. Soc. 2011, 133, 8810–8813. [Google Scholar] [CrossRef]
- Guagnini, F.; Antonik, P.M.; Rennie, M.L.; O’Byrne, P.; Khan, A.R.; Pinalli, R.; Dalcanale, E.; Crowley, P.B. Cucurbit[7]uril-Dimethyllysine Recognition in a Model Protein. Angew. Chem. Int. Ed. 2018, 57, 7126–7130. [Google Scholar] [CrossRef]
- Kumar, K.; Woo, S.M.; Siu, T.; Cortopassi, W.A.; Duarte, F.; Paton, R.S. Cation–π interactions in protein–ligand binding: Theory and data-mining reveal different roles for lysine and arginine. Chem. Sci. 2018, 9, 2655–2665. [Google Scholar] [CrossRef] [PubMed]
- Melegari, M.; Suman, M.; Pirondini, L.; Moiani, D.; Massera, C.; Ugozzoli, F.; Kalenius, E.; Vainiotalo, P.; Mulatier, J.-C.; Dutasta, J.-P.; et al. Supramolecular Sensing with Phosphonate Cavitands. Chem. Eur. J. 2008, 14, 5772–5779. [Google Scholar] [CrossRef] [PubMed]
- Kabsch, W. XDS. Acta Cryst. 2010, D66, 125–132. [Google Scholar] [CrossRef] [PubMed]
- Burla, M.C.; Caliandro, R.; Camalli, M.; Carrozzini, B.; Cascarano, G.L.; Giacovazzo, C.; Mallamo, M.; Mazzone, A.; Polidori, G.; Spagna, R. SIR2011: A new package for crystal structure determination and refinement. J. Appl. Cryst. 2012, 45, 357–361. [Google Scholar] [CrossRef]
- Spek, A.L. PLATON SQUEEZE: A tool for the calculation of the disordered solvent contribution to the calculated structure factors. Acta Cryst. 2015, C71, 9–18. [Google Scholar] [CrossRef]
- Van Dun, S.; Ottmann, C.; Milroy, L.C.; Brunsveld, L. Supramolecular Chemistry Targeting Proteins. J. Am. Chem. Soc. 2017, 139, 13960–13968. [Google Scholar] [CrossRef] [PubMed]
- Menozzi, E.; Busi, M.; Ramingo, R.; Campagnolo, M.; Geremia, S.; Dalcanale, E. Design and Self-Assembly of Ditopic and Tetratopic Cavitand Complexes. Chem. Eur. J. 2005, 11, 3136–3148. [Google Scholar] [CrossRef]
- Busi, M.; Cantadori, B.; Boccini, F.; De Zorzi, R.; Geremia, S.; Dalcanale, E. Molecular Recognition with Ditopic Cavitand Re Complexes. Eur. J. Org. Chem. 2011, 2011, 2629–2642. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compound Tiiii[H, CH3, CH3] is available from the authors. |







| Tiiii(1) H-Bonds | Arg | Lys | His(1) | His(2) |
| N–O2 | 2.71. | 2.71. | 2.81. | 2.71. |
| N–O3 | 2.80 | 2.83 | 2.77 | 2.78 |
| N–OW | 2.74 | 2.72 | 2.64 | 2.66 |
| OW–O1 | 2.90 | 2.87 | 2.76 | 2.81 |
| OW–O4 | 2.83 | 2.90 | 2.73 | 2.87 |
| oop N | 0.70 | 0.55 | 0.89 | 0.89 |
| oop Ow | −0.65 | −0.86 | 0.36 | 0.13 |
| Tiiii(2) H-Bonds | Arg | Lys | His(1) | His(2) |
| O1 | Nη2 2.93 (2.70) | Nζ 2.84 | Nδ1 2.70 | Nδ1 2.69 |
| O2 | Nη1 2.92 (3.47) | Nζ 2.76 | ||
| O3 | Nε 2.76 | Ow 2.68 | Nε2 2.71 | Nδ1 2.67 |
| O4 | Nη2 2.73 (3.45) | Ow 2.71 | ||
| Ow | Nζ 2.72 | |||
| oop | Nη1 −0.54 (−0.64) | Nζ 0.72 | Cε1 −0.47 | Cε1 −0.38 |
| oop | Ow 0.29 |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Brancatelli, G.; Dalcanale, E.; Pinalli, R.; Geremia, S. Probing the Structural Determinants of Amino Acid Recognition: X-Ray Studies of Crystalline Ditopic Host-Guest Complexes of the Positively Charged Amino Acids, Arg, Lys, and His with a Cavitand Molecule. Molecules 2018, 23, 3368. https://doi.org/10.3390/molecules23123368
Brancatelli G, Dalcanale E, Pinalli R, Geremia S. Probing the Structural Determinants of Amino Acid Recognition: X-Ray Studies of Crystalline Ditopic Host-Guest Complexes of the Positively Charged Amino Acids, Arg, Lys, and His with a Cavitand Molecule. Molecules. 2018; 23(12):3368. https://doi.org/10.3390/molecules23123368
Chicago/Turabian StyleBrancatelli, Giovanna, Enrico Dalcanale, Roberta Pinalli, and Silvano Geremia. 2018. "Probing the Structural Determinants of Amino Acid Recognition: X-Ray Studies of Crystalline Ditopic Host-Guest Complexes of the Positively Charged Amino Acids, Arg, Lys, and His with a Cavitand Molecule" Molecules 23, no. 12: 3368. https://doi.org/10.3390/molecules23123368
APA StyleBrancatelli, G., Dalcanale, E., Pinalli, R., & Geremia, S. (2018). Probing the Structural Determinants of Amino Acid Recognition: X-Ray Studies of Crystalline Ditopic Host-Guest Complexes of the Positively Charged Amino Acids, Arg, Lys, and His with a Cavitand Molecule. Molecules, 23(12), 3368. https://doi.org/10.3390/molecules23123368