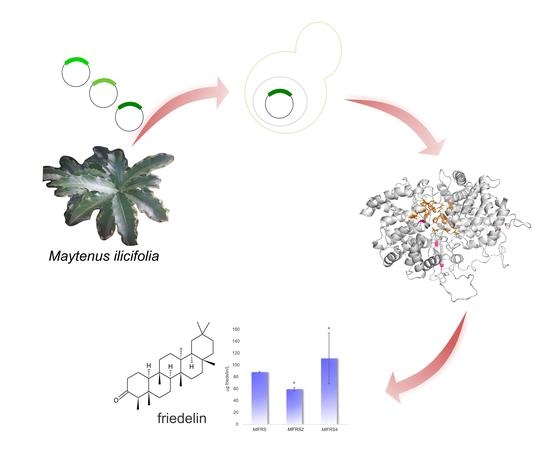
Friedelin in Maytenus ilicifolia Is Produced by Friedelin Synthase Isoforms
Abstract
:1. Introduction
2. Results and Discussion
2.1. There Are Four Coding Sequences for Friedelin Synthase in M. ilicifolia
2.2. MiFRS Gene Characterization and SNPs
2.3. MiFRS NS-SNPs and Enzyme Structure
2.4. Functional Characterization of the Variant Sequences
3. Materials and Methods
3.1. Plant Material
3.2. OSC Cloning
3.3. Phylogenetic Analysis
3.4. MiFRS Gene Sequencing
3.5. In Silico Analysis
3.6. Functional Analysis
3.7. Chemical Analysis and Friedelin Quantification
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Withers, S.; Keasling, J. Biosynthesis and engineering of isoprenoid small molecules. Appl. Microbiol. Biotechnol. 2007, 73, 980–990. [Google Scholar] [CrossRef] [PubMed]
- Wendt, K.U.; Schulz, G.E.; Corey, E.J.; Liu, D.R. Enzyme Mechanisms for Polycyclic Triterpene Formation. Angew. Chem. Int. Ed. Engl. 2000, 39, 2812–2833. [Google Scholar] [CrossRef]
- Xu, R.; Fazio, G.C.; Matsuda, S.P. On the origins of triterpenoid skeletal diversity. Phytochemistry 2004, 65, 261–291. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Yeats, T.; Han, H.; Jetter, R. Cloning and characterization of oxidosqualene cyclases from Kalanchoe daigremontiana: Enzymes catalyzing up to 10 rearrangement steps yielding friedelin and other triterpenoids. J. Biol. Chem. 2010, 285, 29703–29712. [Google Scholar] [CrossRef] [PubMed]
- Thimmappa, R.; Geisler, K.; Louveau, T.; O’Maille, P.; Osbourn, A. Triterpene biosynthesis in plants. Annu. Rev. Plant Biol. 2014, 65, 225–257. [Google Scholar] [CrossRef] [PubMed]
- Rabelo, V.W.; Romeiro, N.C.; Abreu, P.A. Design strategies of oxidosqualene cyclase inhibitors: Targeting the sterol biosynthetic pathway. J. Steroid Biochem. Mol. Biol. 2017, 171, 305–317. [Google Scholar] [CrossRef] [PubMed]
- Benveniste, P. Sterol Metabolism. In The Arabidopsis Book; Biologists TASoP; American Society of Plant Biologists: Rockville, MD, USA, 2002; e-ISSN 1543-8120. [Google Scholar]
- Niero, R.; Faloni de Andrade, S.; Cechinel Filho, V. A Review of the Ethnopharmacology, Phytochemistry and Pharmacology of Plants of the Maytenus Genus. Curr. Pharm. Des. 2011, 17, 1851–1871. [Google Scholar] [CrossRef] [PubMed]
- Antonisamy, P.; Duraipandiyan, V.; Ignacimuthu, S. Anti-inflammatory, analgesic and antipyretic effects of friedelin isolated from Azima tetracantha Lam. in mouse and rat models. J. Pharm. Pharmacol. 2016, 63, 1070–1077. [Google Scholar] [CrossRef] [PubMed]
- Antonisamy, P.; Duraipandiyan, V.; Aravinthan, A.; Al-Dhabi, N.A.; Ignacimuthu, S.; Choi, K.C.; Kim, J.H. Protective effects of friedelin isolated from Azima tetracantha Lam. against ethanol-induced gastric ulcer in rats and possible underlying mechanisms. Eur. J. Pharmacol. 2015, 750, 167–175. [Google Scholar] [CrossRef] [PubMed]
- Pina, E.S.; Silva, D.B.; Teixeira, S.P.; Coppede, J.S.; Furlan, M.; França, S.C.; Lopes, N.P.; Pereira, A.M.S.; Lopes, A.A. Mevalonate-derived quinonemethide triterpenoid from in vitro roots of Peritassa laevigata and their localization in root tissue by MALDI imaging. Sci. Rep. 2016, 6, 22627. [Google Scholar] [CrossRef] [PubMed]
- Paz, T.A.; dos Santos, V.A.; Inácio, M.C.; Dias, N.B.; Palma, M.S.; Pereira, A.M.S.; Furlan, M. Proteome profiling reveals insights into secondary metabolism in Maytenus ilicifolia (Celastraceae) cell cultures producing quinonemethide triterpenes. Plant Cell Tissue Organ Cult. 2017, 130, 405–416. [Google Scholar] [CrossRef]
- Corsino, J.; de Carvalho, P.R.F.; Kato, M.J.; Latorre, L.R.; Oliveira, O.M.M.; Araújo, A.R.; da S Bolzani, V.; França, S.C.; Pereira, A.M.S.; Furlan, M. Biosynthesis of friedelane and quinonemethide triterpenoids is compartmentalized in Maytenus aquifolium and Salacia campestris. Phytochemistry 2000, 55, 741–748. [Google Scholar] [CrossRef]
- Yousef, B.A.; Hassan, H.M.; Zhang, L.Y.; Jiang, Z.Z. Anticancer Potential and Molecular Targets of Pristimerin: A mini-review. Curr. Cancer Drug Targets 2017, 17, 100–108. [Google Scholar] [CrossRef] [PubMed]
- Souza-Moreira, T.M.; Alves, T.B.; Pinheiro, K.A.; Felippe, L.G.; De Lima, G.M.; Watanabe, T.F.; Barbosa, C.C.; Santos, V.A.; Lopes, N.P.; Valentini, S.R.; et al. Friedelin Synthase from Maytenus ilicifolia: Leucine 482 Plays an Essential Role in the Production of the Most Rearranged Pentacyclic Triterpene. Sci. Rep. 2016, 6, 36858. [Google Scholar] [CrossRef] [PubMed]
- Xue, Z.; Duan, L.; Liu, D.; Guo, J.; Ge, S.; Dicks, J.; ÓMáille, P.; Osbourn, A.; Qi, X. Divergent evolution of oxidosqualene cyclases in plants. New Phytol. 2012, 193, 1022–1038. [Google Scholar] [CrossRef] [PubMed]
- Benveniste, P. Biosynthesis and accumulation of sterols. Annu. Rev. Plant Biol. 2004, 55, 429–457. [Google Scholar] [CrossRef] [PubMed]
- Husselstein-Muller, T.; Schaller, H.; Benveniste, P. Molecular cloning and expression in yeast of 2,3-oxidosqualene-triterpenoid cyclases from Arabidopsis thaliana. Plant Mol. Biol. 2001, 45, 75–92. [Google Scholar] [CrossRef] [PubMed]
- Haralampidis, K.; Bryan, G.; Qi, X.; Papadopoulou, K.; Bakht, S.; Melton, R.; Osbourn, A. A new class of oxidosqualene cyclases directs synthesis of antimicrobial phytoprotectants in monocots. Proc. Natl. Acad. Sci. USA 2001, 98, 13431–13436. [Google Scholar] [CrossRef] [PubMed]
- Inagaki, Y.S.; Etherington, G.; Geisler, K.; Field, B.; Dokarry, M.; Ikeda, K.; Mutsukado, Y.; Dicks, J.; Osbourn, A. Investigation of the potential for triterpene synthesis in rice through genome mining and metabolic engineering. New Phytol. 2011, 191, 432–448. [Google Scholar] [CrossRef] [PubMed]
- Carr, I.M.; Robinson, J.I.; Dimitriou, R.; Markham, A.F.; Morgan, A.W.; Bonthron, D.T. Inferring relative proportions of DNA variants from sequencing electropherograms. Bioinformatics 2009, 25, 3244–3250. [Google Scholar] [CrossRef] [PubMed]
- Zang, Y.M.; Li, Y.P.; Qiao, J.; Chen, H.H.; Liu, C.S. Mechanism of genuineness of Glycyrrhiza uralensis based on SNP of β-Amyrin synthase gene. Acta Pharm. Sin. 2015, 50, 906–909. [Google Scholar]
- Shen, Z.; Liu, C.; Wang, X.; Guo, W.; Li, B. Correlation analysis between single nucleotide polymorphism of β-amyrin synthase and content of glycyrrhizic acid in Glycyrrhiza uralensis. China J. Chin. Mater. Med. 2010, 35, 813–816. [Google Scholar]
- Krivoruchko, A.; Nielsen, J. Production of natural products through metabolic engineering of Saccharomyces cerevisiae. Curr. Opin. Biotechnol. 2014, 35, 7–15. [Google Scholar] [CrossRef] [PubMed]
- Corey, E.J.; Matsuda, S.P.; Bartel, B. Molecular cloning, characterization, and overexpression of ERG7, the Saccharomyces cerevisiae gene encoding lanosterol synthase. Proc. Natl. Acad. Sci. USA 1994, 91, 2211–2215. [Google Scholar] [CrossRef] [PubMed]
- Kirby, J.; Romanini, D.W.; Paradise, E.M.; Keasling, J.D. Engineering triterpene production in Saccharomyces cerevisiae—β-Amyrin synthase from Artemisia annua. FEBS J. 2008, 275, 1852–1859. [Google Scholar] [CrossRef] [PubMed]
- Shen, Z.; Liu, C.; Huang, J.; Lin, H.; Su, S. Impacting of polymorphism of β-amyrin synthase from Glycyrrhiza uralensis on its catalytic efficiency. China J. Chin. Mater. Med. 2010, 35, 2941–2944. [Google Scholar]
- Kumar, S.; Stecher, G.; Tamura, K. MEGA7: Molecular Evolutionary Genetics Analysis Version 7.0 for Bigger Datasets. Mol. Biol. Evol. 2016, 33, 1870–1874. [Google Scholar] [CrossRef] [PubMed]
- Saitou, N.; Nei, M. The neighbor-joining method: A new method for reconstructing phylogenetic trees. Mol. Biol. Evol. 1987, 4, 406–425. [Google Scholar] [PubMed]
- Khoomrung, S.; Chumnanpuen, P.; Jansa-Ard, S.; Ståhlman, M.; Nookaew, I.; Borén, J.; Nielsen, J. Rapid quantification of yeast lipid using microwave-assisted total lipid extraction and HPLC-CAD. Anal. Chem. 2013, 85, 4912–4919. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |





| ORF Name | MiFRS (KX147270.1) | MiFRS2 (MG677552) | MiFRS3 (MG677553) | MiFRS4 (MG677554) |
|---|---|---|---|---|
| Nucleotide identity (%) | 99.63 to MiFRS2 and MiFRS3; 99.67 to MiFRS4 | 99.75 to MiFRS3 and 99.79 to MiFRS4 | 99.80 to MiFRS4 | - |
| ORF length (base pairs) | 2316 | 2316 | 2316 | 2316 |
| Amino acid length | 771 | 771 | 771 | 771 |
| Predicted MW (kDa) | 89.16 | 89.16 | 89.17 | 89.17 |
| Predicted pI | 6.04 | 6.24 | 6.17 | 6.17 |
| Protein identity (%) | 99.61 to the other three | 99.74 to MiFRS3 and MiFRS4 | 100 to MiFRS4 | - |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Alves, T.B.; Souza-Moreira, T.M.; Valentini, S.R.; Zanelli, C.F.; Furlan, M. Friedelin in Maytenus ilicifolia Is Produced by Friedelin Synthase Isoforms. Molecules 2018, 23, 700. https://doi.org/10.3390/molecules23030700
Alves TB, Souza-Moreira TM, Valentini SR, Zanelli CF, Furlan M. Friedelin in Maytenus ilicifolia Is Produced by Friedelin Synthase Isoforms. Molecules. 2018; 23(3):700. https://doi.org/10.3390/molecules23030700
Chicago/Turabian StyleAlves, Thaís B., Tatiana M. Souza-Moreira, Sandro R. Valentini, Cleslei F. Zanelli, and Maysa Furlan. 2018. "Friedelin in Maytenus ilicifolia Is Produced by Friedelin Synthase Isoforms" Molecules 23, no. 3: 700. https://doi.org/10.3390/molecules23030700
APA StyleAlves, T. B., Souza-Moreira, T. M., Valentini, S. R., Zanelli, C. F., & Furlan, M. (2018). Friedelin in Maytenus ilicifolia Is Produced by Friedelin Synthase Isoforms. Molecules, 23(3), 700. https://doi.org/10.3390/molecules23030700