Antioxidant Activity of Coconut (Cocos nucifera L.) Protein Fractions
Abstract
:1. Introduction
2. Results and Discussion
2.1. Distribution of Protein Fractions
2.2. SDS-PAGE
2.3. Amino Acid Composition
2.4. Antioxidant Activities
2.4.1. Free Radical Scavenging Activity
2.4.2. Chelating Activity and Reducing Power
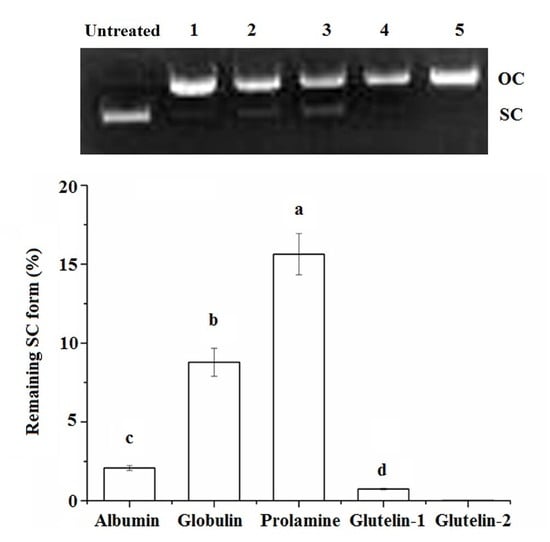
2.4.3. DNA Damage Protection
2.5. LC-MS/MS Analysis
3. Materials and Methods
3.1. Materials
3.2. Fractionation of CCP Fractions
3.3. Sodium Dodecyl Sulfate-Polyacrylamide Gel Electrophoresis (SDS-PAGE)
3.4. Amino Acid Composition
3.5. Antioxidant Activities
3.5.1. ABTS Radical Scavenging Activity
3.5.2. Hydroxyl Radical Scavenging Activity
3.5.3. Superoxide Radical-Scavenging Activity
3.5.4. Metal Chelating Capacity
3.5.5. Reducing Power
3.5.6. Protection on DNA from Oxidative Damage
3.6. LC-MS/MS Analysis
3.7. Statistical Analysis
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Shahidi, F.; Zhong, Y. Novel antioxidants in food quality preservation and health promotion. Eur. J. Lipid Sci. Technol. 2010, 112, 930–940. [Google Scholar] [CrossRef]
- Bhuyan, B.J.; Mugesh, G. Synthesis, characterization and antioxidant activity of angiotensin I converting enzyme inhibitors. Org. Biomol. Chem. 2011, 9, 1356–1365. [Google Scholar] [CrossRef] [PubMed]
- Dong, S.Y.; Zeng, M.Y.; Wang, D.F.; Liu, Z.Y.; Zhao, Y.H.; Yang, H.C. Antioxidant and biochemical properties of protein hydrolysates prepared from Silver carp (Hypophthalmichthys molitrix). Food Chem. 2008, 107, 1483–1493. [Google Scholar] [CrossRef]
- Kwon, K.; Park, K.H.; Rhee, K.C. Fractionation and characterization of proteins from coconut (Cocosnucifera L.). J. Agric. Food Chem. 1996, 44, 1741–1745. [Google Scholar] [CrossRef]
- Angelia, M.R.N.; Garcia, R.N.; Caldo, K.M.P.; Prak, K.; Utsumi, S.; Tecson-Mendoza, E.M. Physicochemical and functional characterization of cocosin, the coconut 11S globulin. Food Sci. Technol. Int. 2010, 16, 225–232. [Google Scholar] [CrossRef]
- Huang, J.M.; Liu, X.Q.; Lan, Q.X.; Lai, X.T.; Luo, Z.; Yang, G.W. Proteomic profile of coconuts. Eur. Food Res. Technol. 2016, 242, 449–455. [Google Scholar] [CrossRef]
- Santana, M.J.; Olivera, A.L.D.; Júnior, L.H.K.Q.; Mandal, S.M.; Matos, C.O.; Dias, R.D. Structural insights into Cn-AMP1, a short disulfide-free multifunctional peptide from green coconut water. FEBS Lett. 2015, 589, 639–644. [Google Scholar] [CrossRef] [PubMed]
- Food and Agricultural Organization; World Health Organization. Protein quality evaluation. In Proceedings of the FAO/WHO Nutrition Meetings, Bethesda, MD, USA, 4–8 December 1989; FAO: Rome, Italy, 1990. [Google Scholar]
- Chee, K.L.; Ling, H.K.; Ayob, M.K. Optimization of trypsin-assisted extraction, physico-chemical characterization, nutritional qualities and functionalities of palm kernel cake protein. LWT Food Sci. Technol. 2012, 46, 419–427. [Google Scholar] [CrossRef]
- Zheng, Y.J.; Li, Y.; Zhang, Y.L.; Zhang, R.G.; Zhang, Q.A.; Zhang, Y.F.; Zhao, S.L. Fractionation, Physicochemical Properties, Nutritional Value, Antioxidant Activity and ACE Inhibition of Palm Kernel Expeller Protein. RSC Adv. 2015, 5, 12613–12623. [Google Scholar] [CrossRef]
- Zhang, M.; Mu, T.H.; Sun, M.J. Purification and identification of antioxidant peptides from sweet potato protein hydrolysates by Alcalase. J. Funct. Foods 2014, 7, 191–200. [Google Scholar] [CrossRef]
- Arts, M.J.T.J.; Dallinga, J.S.; Voss, H.P.; Haenen, G.R.M.M.; Bast, A. A new approach to assess the total antioxidant capacity using the TEAC assay. Food Chem. 2004, 88, 567–570. [Google Scholar] [CrossRef]
- Sarmadi, B.H.; Ismail, A. Antioxidative peptides from food proteins: A review. Peptides 2010, 31, 1949–1956. [Google Scholar] [CrossRef] [PubMed]
- Ren, J.; Zhao, M.; Shi, J.; Wang, J.; Jiang, Y.; Cui, C.; Kakuda, Y.; Xue, S.J. Purification and identification of antioxidant peptides from grass carp muscle hydrolysates by consecutive chromatography and electrospray ionization-mass spectrometry. Food Chem. 2008, 108, 727–736. [Google Scholar] [CrossRef] [PubMed]
- Levent, Y.; Aysun, A.G.; Yusuf, B.; Ahmet, Y. Bioactive, functional and edible film-forming properties of isolated hazelnut (Corylus avellana L.) meal proteins. Food Hydrocoll. 2014, 36, 130–142. [Google Scholar] [CrossRef]
- Jeong, J.B.; De Lumen, B.O.; Jeong, H.J. Lunasin peptide purified from Solanumnigrum L. protects DNA from oxidative damage by suppressing the generation of hydroxyl radical via blocking fenton reaction. Cancer Lett. 2010, 293, 58–64. [Google Scholar] [CrossRef] [PubMed]
- Tapal, A.; Vegarud, G.E.; Sreedhara, A.; Hegde, P.; Inamdarc, S.; Tiku, P.K. In vitro human gastro-intestinal enzyme digestibility of globulin isolate from oil palm (Elaeis guineensis var. tenera) kernel meal and the bioactivity of the digest. RSC Adv. 2016, 6, 20219–20229. [Google Scholar] [CrossRef]
- Zheng, Y.J.; Li, Y.; Zhang, Y.L.; Ruan, X.H.; Zhang, R.G. Purification, characterization, synthesis, in vitro ACE inhibition and in vivo antihypertensive activity of bioactive peptides derived from oil palm kernel glutelin-2 hydrolysates. J. Funct. Food 2017, 28, 48–58. [Google Scholar] [CrossRef]
- Zheng, Y.J.; Li, Y.; Zhang, Y.F. Purification and identification of antioxidativepeptides of palm kernel expeller glutelin-1hydrolysates. RSC Adv. 2017, 7, 54196–54202. [Google Scholar] [CrossRef]
- Gan, R.Y.; Li, H.B.; Gunaratne, A.; Sui, Z.Q.; Corke, H. Effects of fermented edible seeds and their products on human health: Bioactive components and bioactivities. Compr. Rev. Food Sci. Food Saf. 2017, 16, 489–531. [Google Scholar] [CrossRef]
- Bradford, M.M. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein dye binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Laemmli, U.K. Cleavage of structural proteins during the assembly of the head bacteriophage T4. Nature 1970, 227, 680–685. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.S.; Tang, C.H.; Yang, X.Q.; Gao, W.R. Characterization, amino acid composition and in vitro digestibility of hemp (Cannbis sativa L.) proteins. Food Chem. 2008, 107, 11–18. [Google Scholar] [CrossRef]
- Saunders, R.M.; Connor, M.A.; Booth, A.N.; Bickoff, M.M.; Kohler, G.O. Measurement of digestibility of alfalfa protein concentrate by in vivo and in vitro methods. J. Nutr. 1973, 103, 530–535. [Google Scholar] [CrossRef] [PubMed]
- Gao, R.; Yuan, Z.; Zhao, Z.; Gao, X. Mechanism of pyrogallol autoxidation and determination of superoxide dismutase enzyme activity. Bioelectrochem. Bioenerg. 1998, 45, 41–45. [Google Scholar] [CrossRef]
- Yen, G.C.; Chen, H.Y. Antioxidant activity of various tea extracts in relation to their antimutagenicity. J. Agric. Food Chem. 1995, 43, 27–32. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds (coconut cake albumin and globulin) are available from the authors. |




| Amino Acid Residue | Fractions | FAO/WHO | |||||
|---|---|---|---|---|---|---|---|
| Albumin | Globulin | Prolamine | Glutelin-1 | Glutelin-2 | CCPI | ||
| Essential amino acid | |||||||
| Isoleucine | 2.85 ± 0.22 e | 4.12 ± 0.52 b | 5.15 ± 0.34 a | 3.72 ± 0.23 c | 3.22 ± 0.30 d | 4.33 ± 1.70 b | 3.4 |
| Leucine | 4.12 ± 0.36 c | 6.51 ± 0.15 b | 9.55 ± 0.12 a | 6.51 ± 0.17 b | 6.29 ± 0.16 b,c | 6.83 ± 0.78 b | 3.5 |
| Lysine | 5.1 5 ± 0.43 b | 3.53 ± 0.26 d | 8.36 ± 0.39 a | 3.52 ± 0.30 d | 5.54 ± 0.16 b | 4.96 ± 0.65 c | 5.8 |
| Methionine | 1.26 ± 0.25 c | 2.82 ± 0.13 a | 2.95 ± 0.53 a | 2.14 ± 0.31 b | 1.26 ± 0.06 c | 1.57 ± 0.45 c | |
| Phenylalanine | 3.05 ± 0.34 d | 5.86 ±0 .12 a | 5.44 ± 1.54 a | 4.6 5± 0.29 b | 4.45 ± 0.21 c | 4.8 0 ± 0.51 b | |
| Threonine | 3.05 ± 0.35 a | 3.21 ± 0.18 a | 3.14 ± 0.33 a | 3.23 ± 0.04 a | 3.00 ± 0.11 a | 2.73 ± 0.36 b | 2.5 |
| Tryptophan | ND l | ND l | ND l | ND l | ND l | ND l | 2.8 |
| Valine | 3.72 ± 0.33 d | 7.41 ± 0.07 a | 5.95 ± 0.61 b | 4.35 ± 0.57 c | 4.05 ± 0.08 c | 4.80 ± 1.02 c | 6.6 |
| Histidine | 1.74 ± 0.10 c | 2.15 ± 0.35 b | 2.64 ± 0.48 a | 2.54 ± 0.80 a | 1.89 ± 0.11 c | 2.04 ± 0.45 b | 1.9 |
| Tyrosine | 3.35 ± 0.47 b | 3.64 ± 0.10 b | 5.54 ± 0.59 a | 5.68 ± 0.86 a | 3.59 ± 0.72 b | 3.04 ± 1.51 c | 0.5 |
| Nonessential amino acid | |||||||
| Asparagine | 6.95 ± 0.49 b | 8.51 ± 0.42 a | 4.14 ± 0.10 d | 5.7 5± 0.59 c | 8.31 ± 0.50 a | 8.82 ± 0.71 a | |
| Serine | 3.45 ± 0.40 d | 4.55 ± 0.51 b | 5.61 ± 0.31 a | 3.89 ± 0.34 c | 3.84 ± 0.06 c,d | 4.63 ± 1.08 b | |
| Glutamic acid | 23.8 ± 1.56 b | 20.7 ± 4.52 b | 1.92 ± 0.42 d | 17.01 ± 3.61 c | 27.21 ± 0.88 a | 24.50 ± 3.47 b | |
| Glycine | 4.01 ± 0.42 b | 4.78 ± 0.14 a | 2.13 ± 0.57 c | 4.54 ± 0.07 | 4.95 ± 0.31 a | 4.90 ± 0.28 a | |
| Alanine | 2.84 ± 0.24 d | 3.89 ± 0.23 c | 9.21 ± 1.21 a | 3.93 ± 0.45 b | 3.74 ± 0.35 c | 4.06 ± 0.90 b | |
| Arginine | 17.65±0.0.36 a | 15.15± 0.23 b,c | 3.64 ± 0.18 c | 14.25± 1.37 b,c | 16.11 ± 1.27 a | 13.62± 3.27 b,c | |
| Proline | 2.75 ± 0.18 c | 3.14 ± 0.23 b,c | 4.21 ± 0.42 a | 3.22 ± 0.39 b,c | 2.45 ± 0.82 d | 3.60 ± 0.50 b | |
| Bigelow parameters | |||||||
| NPS f | 0.22 ± 0.03 d | 0.31 ± 0.06 b | 0.45 ± 0.04 a | 0.32 ± 0.03 b | 0.24 ± 0.11 d | 0.28 ± 0.05 c | |
| P g | 2.79 ± 0.44 a | 1.63 ± 0.03 b | 0.58 ± 67 c | 1.53 ± 0.16 b,c | 2.46 ± 0.13 a,b | 1.97 ± 0.31 b | |
| HФ h | 844.00 e | 1100.01 a | 1013.29 c | 990.21 d | 1111.65 a | 1084.66 b | |
| Nutritional quality | |||||||
| TEAA i | 28.29 ± 1.19 c | 39.25 ± 0.95 b | 48.72 ± 2.30 a | 36.34 ± 1.41 b | 33.29±1.12 b,c | 35.10 ± 3.15 b | 12.7 |
| E/T j (%) | 31.52 d | 39.26 c | 61.22 a | 40.86 b | 33.32 d | 35.37 c | |
| IVPD k (g/100 g) | 90.31 ± 3.48 a | 88.26 ± 2.58 b | ND l | 72.74 ± 4.11 c,d | 76.35 ± 5.12 c | 86.97 ± 1.62 a | |
| Fractions | Free Radical Scavenging Ability (%) a | Antioxidant Activity | |||
|---|---|---|---|---|---|
| ABTS Radical | Hydroxyl Radical | Superoxide Radical | Chelating Activity (%) | Reducing Power | |
| Albumin | 5.04 ± 0.59 d,e | 8.83 ± 0.98 f | 14.99 ± 2.10 e | 5.13 ± 1.44 e,f | 0.121 ± 0.010 c |
| Globulin | 2.07 ± 0.64 e | 16.87 ± 1.46 e | 10.91 ± 0.79 e | 89.14 ± 3.42 b | 0.016 ± 0.005 e |
| Prolamine | 66.05 ± 1.17 b | 68.27 ± 6.52 c | 56.31 ± 2.60 c | 80.38 ± 6.11 c | 0.156 ± 0.003 b |
| Glutelin-1 | 52.58 ± 0.22 c | 20.48 ± 1.85 d | 67.82 ± 2.72 b | 3.60 ± 0.48 f | 0.138 ± 0.003 b |
| Glutelin-2 | 53.76 ± 0.25 c | 68.27 ±2.55 c | 4.14 ± 0.75 f | 5.02 ± 1.17 e,f | 0.100 ± 0.003 d |
| BHT g | 5.81 ± 1.46 d,e | 77.54 ± 3.45 b | 48.68 ± 0.83 d | 41.45 ± 0.78 d | 0.002 ± 0.000 f |
| Fraction | Peptide Sequence a | Molecular Weight (Da) | Possible Biological Activities | References |
|---|---|---|---|---|
| Globulin | NFLEK | 649.34 | ||
| ATHELR | 725.38 | |||
| EDKLER | 788.4 | |||
| AGTIVSFANR | 1034.55 | Antioxidative | [17] | |
| SWPFGESR | 964.44 | |||
| RPFNLFHK | 1057.58 | Antioxidative | [17] | |
| GREEEEGR | 960.43 | |||
| TWLAGR | 702.38 | |||
| Glutelin-2 | VIEPR | 612.36 | Antioxidative | [18] |
| RVIEPR | 768.46 | Antioxidative | [17] | |
| QFLLAGR | 803.46 | |||
| ENILR | 643.36 | |||
| KLQCR | 703.38 | |||
| IKQNIGDPR | 1039.58 | |||
| QNIGDPR | 798.40 | |||
| RADVFNPR | 973.51 | Antioxidative | [17] | |
| ADVFNPR | 817.41 | Antioxidative | [17,19] | |
| ITTLNSEK | 904.49 | |||
| LPILR | 610.45 | Antioxidative | [18] | |
| VVLYR | 648.40 | Antioxidative | [18] |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, Y.; Zheng, Y.; Zhang, Y.; Xu, J.; Gao, G. Antioxidant Activity of Coconut (Cocos nucifera L.) Protein Fractions. Molecules 2018, 23, 707. https://doi.org/10.3390/molecules23030707
Li Y, Zheng Y, Zhang Y, Xu J, Gao G. Antioxidant Activity of Coconut (Cocos nucifera L.) Protein Fractions. Molecules. 2018; 23(3):707. https://doi.org/10.3390/molecules23030707
Chicago/Turabian StyleLi, Yan, Yajun Zheng, Yufeng Zhang, Jianguo Xu, and Gang Gao. 2018. "Antioxidant Activity of Coconut (Cocos nucifera L.) Protein Fractions" Molecules 23, no. 3: 707. https://doi.org/10.3390/molecules23030707
APA StyleLi, Y., Zheng, Y., Zhang, Y., Xu, J., & Gao, G. (2018). Antioxidant Activity of Coconut (Cocos nucifera L.) Protein Fractions. Molecules, 23(3), 707. https://doi.org/10.3390/molecules23030707