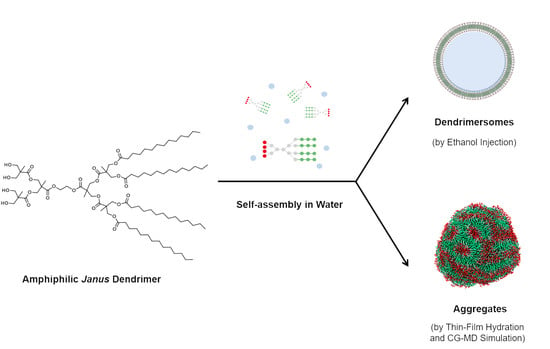
Self-Assembly Behavior of Amphiphilic Janus Dendrimers in Water: A Combined Experimental and Coarse-Grained Molecular Dynamics Simulation Approach
Abstract
:1. Introduction
2. Results
2.1. Formation and Characterization of Assemblies by Experimental Methods
2.1.1. Giant Assemblies
2.1.2. Small Assemblies
2.2. Coarse-Grained Molecular Dynamic Simulation
3. Discussion
4. Materials and Methods
4.1. General Information
4.2. Instruments for Dendrimers Characterization
4.2.1. Nuclear Magnetic Resonance (NMR)
4.2.2. Mass Spectra
4.3. Synthesis and Characterization of Amphiphilic Janus Dendrimers
4.3.1. Dendron 1 and General Esterification Procedure
4.3.2. Dendron 2
4.3.3. Dendron 3
4.3.4. Amphiphilic Janus Dendrimer (JD)
4.4. Formation and Characterization of Assemblies (Experimental Method)
4.4.1. Giant Assemblies
4.4.2. Small Assemblies
4.5. Coarse-Grained Molecular Dynamic Simulation
4.6. Statistics
5. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kalhapure, R.S.; Kathiravan, M.K.; Akamanchi, K.G.; Govender, T. Dendrimers—From organic synthesis to pharmaceutical applications: An update. Pharm. Dev. Technol. 2013, 20, 22–40. [Google Scholar] [CrossRef] [PubMed]
- Fedeli, E.; Lancelot, A.; Serrano, J.L.; Calvo, P.; Sierra, T. Self-assembling amphiphilic Janus dendrimers: Mesomorphic properties and aggregation in water. New J. Chem. 2015, 39, 1960–1967. [Google Scholar] [CrossRef]
- Percec, V.; Wilson, D.A.; Leowanawat, P.; Wilson, C.J.; Hughes, A.D.; Kaucher, M.S.; Hammer, D.A.; Levine, D.H.; Kim, A.J.; Bates, F.S.; et al. Self-assembly of Janus dendrimers into uniform dendrimersomes and other complex architectures. Science 2010, 328, 1009–1014. [Google Scholar] [CrossRef] [PubMed]
- Sikwal, D.R.; Kalhapure, R.S.; Govender, T. An emerging class of amphiphilic dendrimers for pharmaceutical and biomedical applications: Janus amphiphilic dendrimers. Eur. J. Pharm. Sci. 2017, 97, 113–134. [Google Scholar] [CrossRef] [PubMed]
- Filippi, M.; Patrucco, D.; Martinelli, J.; Botta, M.; Castro-Hartmann, P.; Tei, L.; Terreno, E. Novel stable dendrimersome formulation for safe bioimaging applications. Nanoscale 2015, 7, 12943–12954. [Google Scholar] [CrossRef] [PubMed]
- Filippi, M.; Martinelli, J.; Mulas, G.; Ferraretto, M.; Teirlinck, E.; Botta, M.; Tei, L.; Terreno, E. Dendrimersomes: A new vesicular nano-platform for MR-molecular imaging applications. Chem. Commun. 2014, 50, 3453–3456. [Google Scholar] [CrossRef] [PubMed]
- Filippi, M.; Catanzaro, V.; Patrucco, D.; Botta, M.; Tei, L.; Terreno, E. First in vivo MRI study on theranostic dendrimersomes. J. Control. Release 2017, 248, 45–52. [Google Scholar] [CrossRef] [PubMed]
- Nazemi, A.; Gillies, E.R. Dendrimersomes with photodegradable membranes for triggered release of hydrophilic and hydrophobic cargo. Chem. Commun. 2014, 50, 11122–11125. [Google Scholar] [CrossRef] [PubMed]
- Peterca, M.; Percec, V.; Leowanawat, P.; Bertin, A. Predicting the size and properties of dendrimersomes from the lamellar structure of their amphiphilic Janus dendrimers. J. Am. Chem. Soc. 2011, 133, 20507–20520. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, H.J.; Hughes, A.D.; Moussodia, R.O.; Bertin, A.; Chen, Y.; Pochan, D.J.; Heiney, P.A.; Klein, M.L.; Percec, V. Self-assembly of amphiphilic Janus dendrimers into uniform onion-like dendrimersomes with predictable size and number of bilayers. Proc. Natl. Acad. Sci. USA 2014, 111, 9058–9063. [Google Scholar] [CrossRef] [PubMed]
- Zhang, S.; Sun, H.J.; Hughes, A.D.; Draghici, B.; Lejnieks, J.; Leowanawat, P.; Bertin, A.; Otero De Leon, L.; Kulikov, O.V.; Chen, Y.; et al. “Single-Single” Amphiphilic Janus Dendrimers Self-Assemble into Uniform Dendrimersomes with Predictable Size. ACS Nano 2014, 8, 1554–1565. [Google Scholar] [CrossRef] [PubMed]
- Wang, P.; Ma, Y.; Liu, Z.; Yan, Y.; Sun, X.; Zhang, J. Vesicle formation of catanionic mixtures of CTAC/SDS induced by ratio: A coarse-grained molecular dynamic simulation study. RSC Adv. 2016, 6, 13442–13449. [Google Scholar] [CrossRef]
- Marrink, S.J.; de Vries, A.H.; Mark, A.E. Coarse Grained Model for Semiquantitative Lipid Simulations. J. Phys. Chem. B 2004, 108, 750–760. [Google Scholar] [CrossRef]
- Thota, N.; Luo, Z.; Hu, Z.; Jiang, J. Self-Assembly of Amphiphilic Peptide (AF)6H5K15: Coarse-Grained Molecular Dynamics Simulation. J. Phys. Chem. B 2013, 117, 9690–9698. [Google Scholar] [CrossRef] [PubMed]
- Márquez-Miranda, V.; Araya-Durán, I.; Camarada, M.B.; Comer, J.; Valencia-Gallegos, J.A.; González-Nilo, F.D. Self-Assembly of Amphiphilic Dendrimers: The Role of Generation and Alkyl Chain Length in siRNA Interaction. Sci. Rep. 2016, 6, 29436. [Google Scholar] [CrossRef] [PubMed]
- Nummelin, S.; Selin, M.; Legrand, S.; Ropponen, J.; Seitsonen, J.; Nykänen, A.; Koivisto, J.; Hirvonen, J.; Kostiainen, M.A.; Bimbo, L.M. Modular synthesis of self-assembling Janus-dendrimers and facile preparation of drug-loaded dendrimersomes. Nanoscale 2017, 9, 7189–7198. [Google Scholar] [CrossRef] [PubMed]
- Tsumoto, K.; Matsuo, H.; Tomita, M.; Yoshimura, T. Efficient formation of giant liposomes through the gentle hydration of phosphatidylcholine films doped with sugar. Colloids Surf. B Biointerfaces 2009, 68, 98–105. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez, N.; Pincet, F.; Cribier, S. Giant vesicles formed by gentle hydration and electroformation: A comparison by fluorescence microscopy. Colloids Surf. B Biointerfaces 2005, 42, 125–130. [Google Scholar] [CrossRef] [PubMed]
- Ianiro, A.; Patterson, J.; González García, A.; van Rijt, M.M.J.; Hendrix, M.M.R.M.; Sommerdijk, N.A.J.M.; Voets, I.K.; Esteves, A.C.C.; Tuinier, R. A roadmap for poly(ethylene oxide)-block-poly-ε-caprolactone self-assembly in water: Prediction, synthesis, and characterization. J. Polym. Sci. Part B Polym. Phys. 2018, 56, 330–339. [Google Scholar] [CrossRef]
- Ruozi, B.; Belletti, D.; Tombesi, A.; Tosi, G.; Bondioli, L.; Forni, F.; Vandelli, M.A. AFM, ESEM, TEM, and CLSM in liposomal characterization: A comparative study. Int. J. Nanomed. 2011, 6, 557–563. [Google Scholar] [CrossRef] [PubMed]
- Antonietti, M.; Förster, S. Vesicles and Liposomes: A Self-Assembly Principle Beyond Lipids. Adv. Mater. 2003, 15, 1323–1333. [Google Scholar] [CrossRef]
- Giustini, M.; Bellinazzo, C.; Galantini, L.; Mallardi, A.; Palazzo, G.; Sennato, S.; Bordi, F.; Rissanen, K. Incorporation of the bacterial reaction centre into dendrimersomes. Colloids Surf. A Physicochem. Eng. Asp. 2012, 413, 38–43. [Google Scholar] [CrossRef]
- Luman, N.R.; Grinstaff, M.W. Synthesis and aqueous aggregation properties of amphiphilic surface-block dendrimers. Org. Lett. 2005, 7, 4863–4866. [Google Scholar] [CrossRef] [PubMed]
- Ruozi, B.; Tosi, G.; Tonelli, M.; Bondioli, L.; Mucci, A.; Forni, F.; Vandelli, M.A. AFM phase imaging of soft-hydrated samples: A versatile tool to complete the chemical-physical study of liposomes. J. Liposome Res. 2009, 19, 59–67. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bibi, S.; Kaur, R.; Henriksen-Lacey, M.; McNeil, S.E.; Wilkhu, J.; Lattmann, E.; Christensen, D.; Mohammed, A.R.; Perrie, Y. Microscopy imaging of liposomes: From coverslips to environmental SEM. Int. J. Pharm. 2011, 417, 138–150. [Google Scholar] [CrossRef] [PubMed]
- Parent, L.R.; Bakalis, E.; Ramírez-Hernández, A.; Kammeyer, J.K.; Park, C.; De Pablo, J.; Zerbetto, F.; Patterson, J.P.; Gianneschi, N.C. Directly Observing Micelle Fusion and Growth in Solution by Liquid-Cell Transmission Electron Microscopy. J. Am. Chem. Soc. 2017, 139, 17140–17151. [Google Scholar] [CrossRef] [PubMed]
- Liao, H.-G.; Zheng, H. Liquid Cell Transmission Electron Microscopy. Annu. Rev. Phys. Chem. 2016, 67, 719–747. [Google Scholar] [CrossRef] [PubMed]
- Han, M.; Hong, M.; Sim, E. Influence of the block hydrophilicity of AB2 miktoarm star copolymers on cluster formation in solutions. J. Chem. Phys. 2011, 134, 204901. [Google Scholar] [CrossRef] [PubMed]
- Yamamoto, S.; Maruyama, Y.; Hyodo, S. Dissipative particle dynamics study of spontaneous vesicle formation of amphiphilic molecules. J. Chem. Phys. 2002, 116, 5842. [Google Scholar] [CrossRef]
- Wang, Z.; He, X. Dynamics of vesicle formation from lipid droplets: Mechanism and controllability. J. Chem. Phys. 2009, 130, 94905. [Google Scholar] [CrossRef] [PubMed]
- Shinoda, W.; Devane, R.; Klein, M.L. Zwitterionic lipid assemblies: Molecular dynamics studies of monolayers, bilayers, and vesicles using a new coarse grain force field. J. Phys. Chem. B 2010, 114, 6836–6849. [Google Scholar] [CrossRef] [PubMed]
- Israelachvili, J.N.; Mitchell, D.J.; Ninham, B.W. Theory of self-assembly of hydrocarbon amphiphiles into micelles and bilayers. J. Chem. Soc. Faraday Trans. 2 1976, 72, 1525. [Google Scholar] [CrossRef]
- Arai, N.; Yoshimoto, Y.; Yasuoka, K.; Ebisuzaki, T. Self-assembly behaviours of primitive and modern lipid membrane solutions: A coarse-grained molecular simulation study. Phys. Chem. Chem. Phys. 2016, 18, 19426–19432. [Google Scholar] [CrossRef] [PubMed]
- Kremer, J.M.; Esker, M.W.; Pathmamanoharan, C.; Wiersema, P.H. Vesicles of Variable Diameter Prepared by a Modified Injection Method. Biochemistry 1977, 16, 3932–3935. [Google Scholar] [CrossRef] [PubMed]
- Garcia-Manrique, P.; Matos, M.; Gutierrez, G.; Estupiñan, O.R.; Blanco-Lopez, M.C.; Pazos, C. Using Factorial Experimental Design to Prepare Size-Tuned Nanovesicles. Ind. Eng. Chem. Res. 2016, 55, 9164–9175. [Google Scholar] [CrossRef]
- Moore, J.S.; Stupp, S.I. Room Temperature Polyesterification. Macromolecules 1990, 23, 65–70. [Google Scholar] [CrossRef]
- Appel, R.; Fuchs, J.; Tyrrell, S.M.; Korevaar, P.A.; Stuart, M.C.A.; Voets, I.K.; Schönhoff, M.; Besenius, P. Steric Constraints Induced Frustrated Growth of Supramolecular Nanorods in Water. Chem. A Eur. J. 2015, 21, 19257–19264. [Google Scholar] [CrossRef] [PubMed]
- Ihre, H.; Hult, A.; Fréchet, J.M.J.; Gitsov, I. Double-Stage Convergent Approach for the Synthesis of Functionalized Dendritic Aliphatic Polyesters Based on 2,2-Bis(hydroxymethyl)propionic Acid. Macromolecules 1998, 31, 4061–4068. [Google Scholar] [CrossRef]
- Tuuttila, T.; Lahtinen, M.; Huuskonen, J.; Rissanen, K. Synthesis and thermal behavior of Janus dendrimers, part 2. Thermochim. Acta 2010, 497, 109–116. [Google Scholar] [CrossRef]
- Walde, P.; Cosentino, K.; Engel, H.; Stano, P. Giant vesicles: Preparations and applications. ChemBioChem 2010, 11, 848–865. [Google Scholar] [CrossRef] [PubMed]
- Meure, L.A.; Foster, N.R.; Dehghani, F. Conventional and dense gas techniques for the production of liposomes: A review. AAPS PharmSciTech 2008, 9, 798–809. [Google Scholar] [CrossRef] [PubMed]
- Gentine, P.; Bourel-Bonnet, L.; Frisch, B. Modified and derived ethanol injection toward liposomes: Development of the process. J. Liposome Res. 2013, 23, 11–19. [Google Scholar] [CrossRef] [PubMed]
- Schindelin, J.; Arganda-Carreras, I.; Frise, E.; Kaynig, V.; Longair, M.; Pietzsch, T.; Preibisch, S.; Rueden, C.; Saalfeld, S.; Schmid, B.; et al. Fiji: An open-source platform for biological-image analysis. Nat. Methods 2012, 9, 676–682. [Google Scholar] [CrossRef] [PubMed]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef] [PubMed]
- Periole, X.; Marrink, S.-J. The Martini Coarse-Grained Force Field. In Biomolecular Simulations. Methods in Molecular Biology (Methods and Protocols); Monticelli, L., Salonen, E., Eds.; Humana Press: Totowa, NJ, USA, 2013; Volume 924, pp. 533–565. ISBN 978-1-62703-016-8. [Google Scholar]
- Bradley, R.; Radhakrishnan, R. Coarse-grained models for protein-cell membrane interactions. Polymers 2013, 5, 890–936. [Google Scholar] [CrossRef] [PubMed]
- Bussi, G.; Donadio, D.; Parrinello, M. Canonical sampling through velocity rescaling. J. Chem. Phys. 2007, 126, 14101. [Google Scholar] [CrossRef] [PubMed]
- Thota, N.; Jiang, J. Self-assembly of amphiphilic peptide (AF)6H5K 15 derivatives: Roles of hydrophilic and hydrophobic residues. J. Phys. Chem. B 2014, 118, 2683–2692. [Google Scholar] [CrossRef] [PubMed]
- Essmann, U.; Perera, L.; Berkowitz, M.L.; Darden, T.; Lee, H.; Pedersen, L.G. A smooth particle mesh Ewald method. J. Chem. Phys. 1995, 103, 8577–8593. [Google Scholar] [CrossRef]
- Humphrey, W.; Dalke, A.; Schulten, K. VMD: Visual molecular dynamics. J. Mol. Graph. 1996, 14, 33–38. [Google Scholar] [CrossRef]
Sample Availability: Sample of the dendrimer are available from the authors. |








© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Elizondo-García, M.E.; Márquez-Miranda, V.; Araya-Durán, I.; Valencia-Gallegos, J.A.; González-Nilo, F.D. Self-Assembly Behavior of Amphiphilic Janus Dendrimers in Water: A Combined Experimental and Coarse-Grained Molecular Dynamics Simulation Approach. Molecules 2018, 23, 969. https://doi.org/10.3390/molecules23040969
Elizondo-García ME, Márquez-Miranda V, Araya-Durán I, Valencia-Gallegos JA, González-Nilo FD. Self-Assembly Behavior of Amphiphilic Janus Dendrimers in Water: A Combined Experimental and Coarse-Grained Molecular Dynamics Simulation Approach. Molecules. 2018; 23(4):969. https://doi.org/10.3390/molecules23040969
Chicago/Turabian StyleElizondo-García, Mariana E., Valeria Márquez-Miranda, Ingrid Araya-Durán, Jesús A. Valencia-Gallegos, and Fernando D. González-Nilo. 2018. "Self-Assembly Behavior of Amphiphilic Janus Dendrimers in Water: A Combined Experimental and Coarse-Grained Molecular Dynamics Simulation Approach" Molecules 23, no. 4: 969. https://doi.org/10.3390/molecules23040969
APA StyleElizondo-García, M. E., Márquez-Miranda, V., Araya-Durán, I., Valencia-Gallegos, J. A., & González-Nilo, F. D. (2018). Self-Assembly Behavior of Amphiphilic Janus Dendrimers in Water: A Combined Experimental and Coarse-Grained Molecular Dynamics Simulation Approach. Molecules, 23(4), 969. https://doi.org/10.3390/molecules23040969