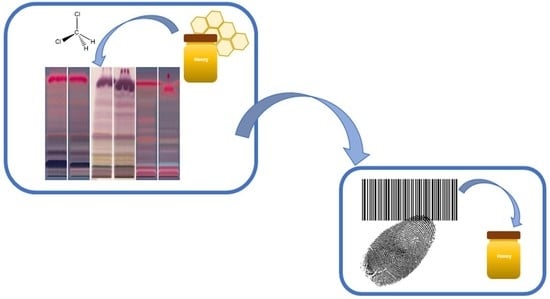
HPTLC Fingerprinting—Rapid Method for the Differentiation of Honeys of Different Botanical Origin Based on the Composition of the Lipophilic Fractions
Abstract
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. Reagents and Materials
3.2. Honey Samples and Melissopalynological Analyses
3.3. Extraction of Lipophilic Fraction from Honeys
3.3.1. Ultrasound Assisted Extraction (UAE)
3.3.2. Solid Phase Extraction (SPE)
3.4. HPTLC Separation of Honey Extracts
Derivatisation
3.5. Statistical Analysis
4. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Bodganov, S.; Jurendic, T.; Sieber, R.; Gallmann, P. Honey for nutrition and health a review. J. Agric. Food. Chem. 2008, 276, 677–689. [Google Scholar]
- Nagai, T.; Inoue, R.; Kanamori, N.; Suzuki, N.; Nagashima, T. Characterization of honey from different floral sources. Its functional properties and effects of honey species on storage of meat. Food Chem. 2006, 97, 256–262. [Google Scholar] [CrossRef]
- Ajibola, A.; Chamunorwa, J.P.; Erlwanger, K.H. Nutraceutical values of natural honey and its contribution to human health and wealth. Nutr. Metab. 2012, 9, 61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Forrest, R.D. Early history of wound treatment. J. R. Soc. Med. 1982, 75, 198–205. [Google Scholar] [PubMed]
- Da Silva Priscila, M.; Gauche, C.; Valdemiro, L.; Costa, A.C.O.; Fett, R. Honey: Chemical composition, stability and authenticity. Food Chem. 2016, 196, 309–323. [Google Scholar] [CrossRef] [PubMed]
- Escuredo, O.; Míguez, M.; Fernández-González, M.; Seijo, M.C. Nutritional value and antioxidant activity of honeys produced in a European Atlantic area. Food Chem. 2013, 138, 851–856. [Google Scholar] [CrossRef] [PubMed]
- Tornuk, F.; Karaman, S.; Ozturk, I.; Toker, O.S.; Tastemur, B.; Sagdic, O.; Dogan, M.; Kayacier, A. Quality characterization of artisanal and retail Turkish blossom honeys: Determination of physicochemical; microbiological; bioactive properties and aroma profile. Ind. Crops. Prod. 2013, 46, 124–131. [Google Scholar] [CrossRef]
- Soares, S.; Amaral, J.S.; Oliveira, M.B.P.P.; Mafra, I.A. Comprehensive Review on the Main Honey Authentication Issues: Production and Origin. Compr. Rev. Food Sci. Food Saf. 2017, 16, 1072–1100. [Google Scholar] [CrossRef]
- Wang, A.; Guo, Q.; Wang, L.; Lin, L.; Shi, H.; Cao, H.; Cao, B. Detection of honey adulteration with starch syrup by high performance liquid chromatography. Food Chem. 2015, 172, 669–674. [Google Scholar] [CrossRef] [PubMed]
- Anklam, E. A review of the analytical methods to determine the geographical and botanical origin of honey. Food Chem. 1998, 63, 549–562. [Google Scholar] [CrossRef]
- Louveaux, J.; Maurizio, A.; Vorwohl, G. Methods of melissopalinology. Bee World 1978, 59, 139–157. [Google Scholar] [CrossRef]
- Kukurová, K.; Karovičová, J.; Kohajdová, Z.; Bíliková, K. Authentication of honey by multivariate analysis of its physico–chemical parameters. J. Food. Nutr. Res. 2008, 47, 170–180. [Google Scholar]
- Jasicka-Misiak, I.; Makowicz, E.; Stanek, N. Polish Yellow Sweet Clover Melilotus officinalis L. Honey, Chromatographic Fingerprints, and Chemical Markers. Molecules 2017, 22, 138. [Google Scholar] [CrossRef] [PubMed]
- Kaskoniene, V.; Venskutonis, P.R. Floral markers in honey of various botanical and geographic origins: A review. Compr. Rev. Food Sci. Food Saf. 2010, 9, 620–634. [Google Scholar] [CrossRef]
- Jerković, I.; Marijanović, Z.; Staver, M.M. Screening of natural organic volatiles from Prunus mahaleb L. Honey: Coumarin and vomifoliol as nonspecific biomarkers. Molecules 2011, 16, 2507–2518. [Google Scholar] [CrossRef] [PubMed]
- Zieliński, Ł.; Deja, S.; Jasicka-Misiak, I.; Kafarski, P. Chemometrics as a Tool of Origin Determination of Polish Monofloral and Multifloral Honeys. J. Agric. Food Chem. 2014, 6213, 2973–2981. [Google Scholar] [CrossRef] [PubMed]
- Jerković, I.; Kuś, P.M. Terpenes in honey: Occurrence; origin and their role as chemical biomarkers. RSC Adv. 2014, 4, 31710–31728. [Google Scholar] [CrossRef]
- Ciulu, M.; Spano, N.; Pilo, M.I.; Sanna, G. Recent Advances in the Analysis of Phenolic Compounds in Unifloral Honeys. Molecules 2016, 21, 451. [Google Scholar] [CrossRef] [PubMed]
- Odeh, I.; Abu-Lafi, S.; Dewik, H.; Al-Najjar, I.; Imam, A.; Dembitsky, V.M.; Hanuš, L.O. A variety of volatile compoundsas markers in Palestinian honey from Thymus capitatus, Thymelaea hirsute and Tolpis virgate. Food Chem. 2007, 101, 1393–1397. [Google Scholar] [CrossRef]
- Piasenzotto, L.; Gracco, L.; Conte, L. Solid phase micro extraction SPME applied to honey quality control. J. Agric. Food Chem. 2003, 83, 1037–1044. [Google Scholar] [CrossRef]
- Tanner, G.; Czerwenka, C. LC-MS/MS Analysis of Neonicotinoid Insecticides in Honey: Methodology and Residue Findings in Austrian Honeys. Compr. Rev. Food Sci. Food Saf. 2011, 5923, 12271–12277. [Google Scholar] [CrossRef] [PubMed]
- Azevedo, M.S.; Seraglio, S.K.T.; Rocha, G.; Balderas, C.B.; Piovezan, M.; Gonzaga, L.V.; de Barcellos Falkenberg, D.; Fett, R.; de Oliveira, M.A.L.; Costa, A.C.O. Free amino acid determination by GC-MS combined with a chemometric approach for geographical classification of bracatinga honeydew honey (Mimosa scabrella Bentham). Food Control 2017, 78, 383–392. [Google Scholar] [CrossRef]
- Jandrić, Z.; Frew, R.D.; Fernandez-Cedi, L.N.; Cannavan, A. An investigative study on discrimination of honey of various floral and geographical origins using UPLC-QToF MS and multivariate data analysis. Food Control 2017, 72, 189–197. [Google Scholar] [CrossRef]
- Svečnjak, L.; Prđun, S.; Rogina, J.; Bubalo, D.; Jerković, I. Characterization of Satsuma mandarin Citrus unshiu Marc. nectar-to-honey transformation pathway using FTIR-ATR spectroscopy. Food Chem. 2017, 232, 286–294. [Google Scholar] [CrossRef] [PubMed]
- Kasprzyk, J.; Depciuch, J.; Grabek-Lejko, D.; Parlinska-Wojtan, M. FTIR-ATR spectroscopy of pollen and honey as a tool for unifloral honey authentication. The case study of rape honey. Food Control. 2018, 84, 33–40. [Google Scholar] [CrossRef]
- Guelpa, A.; Marini, F.; Plessis, A.; Slabbert, R.; Manley, M. Verification of authenticity and fraud detection in South African honey using NIR spectroscopy. Food Control 2017, 73, 1388–1396. [Google Scholar] [CrossRef]
- Beitlich, N.; Lübken, T.; Kaiser, M.; Ispiryan, L.; Speer, K. Fluorescent Pteridine Derivatives as New Markers for the Characterization of Genuine Monofloral New Zealand Manuka Leptospermum scoparium Honey. J. Agric. Food Chem. 2016, 6446, 8886–8891. [Google Scholar] [CrossRef] [PubMed]
- Locher, C.; Neumann, J.; Sostaric, T. Authentication of Honeys of Different Floral Origins via High-Performance Thin-Layer Chromatographic Fingerprinting. J. Planar. Chromatogr. 2017, 30, 57–62. [Google Scholar] [CrossRef]
- Rizelio, V.M.; Tenfen, L.; De Silveira, R.; Gonzaga, L.V.; Carolina, A.; Costa, O.; Fett, R. Development of a fast capillary electrophoresis method for determination of carbohydrates in honey samples. Talanta 2012, 93, 62–66. [Google Scholar] [CrossRef] [PubMed]
- Da Silva, V.L.; Cerqueira, M.R.F.; Lowinsohn, D.; Matos, M.A.C.; Matos, R.C. Amperometric detection of ascorbic acid in honey using ascorbate oxidase immobilised on amberlite IRA-743. Food Chem. 2012, 133, 1050–1054. [Google Scholar] [CrossRef]
- Buldini, P.L.; Cavalli, S.; Mevoli, A.; Sharma, J.L. Ion chromatographic and voltammetric determination of heavy and transition metals in honey. Food Chem. 2011, 73, 487–495. [Google Scholar] [CrossRef]
- Centi, S.; Stoica, A.I.; Laschi, S.; Mascini, M. Development of an electrochemical immunoassay based on the use of an eight-electrodes screen-printed array coupled with magnetic beads for the detection of antimicrobial sulfonamides in honey. Electroanal 2010, 22, 1881–1888. [Google Scholar] [CrossRef]
- Stanek, N.; Jasicka-Misiak, I. HPTLC Phenolic Profiles as Useful Tools for the Authentication of Honey. Food Anal. Method. 2018. [Google Scholar] [CrossRef]
Sample Availability: The honey samples are available from the authors for limited time. |





| Honey | Derivatised UV | 366 nm Derivatised | 254 nm Derivatised | Derivatised UV | 366 nm Derivatised | 254 nm Derivatised | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UAE | SPE | |||||||||||
| *Rf | Zone Color | *Rf | Zone Color | *Rf | Zone Color | *Rf | Zone Color | *Rf | Zone Color | *Rf | Zone Color | |
| Buckwheat (cv.KORA) (Fagopyrum esculentum Moench) | 0.35 0.23 0.11 | weak violet brown deep green | 0.45 0.23 0.11 | weak green brown deep brown | 0.23 0.34 0.45 0.11 | brown orange weak green deep brown | 0.05 0.28 0.08 | weak orange pink weak brown | 0.60 0.42 0.33 0.09 | blue weak gray blue orange | 0.08 0.18 0.33 0.43 | orange deep orange weak brown weak green |
| Buckwheat (cv.PANDA) (Fagopyrum esculentum Moench) | 0.35 0.23 0.11 0.05 | weak violet brown deep green pink | 0.45 0.23 0.11 0.14 0.05 | weak green brown deep brown light blue pink | 0.14 0.05 0.11 0.23 0.45 | weak blue pink deep brown brown weak green | 0.08 0.04 0.19 0.28 | weak brown violet weak brown pink | 0.42 0.33 0.12 0.18 0.08 0.05 | weak gray blue blue brown orange pink | 0.08 0.18 0.12 0.33 0.43 | orange deep orange blue weak brown weak green |
| Raspberry (Rubus spp.) | 0.11 0.19 0.22 0.33 | deep black weak brown gray green | 0.11 0.13 0.19 0.33 | deep black deep pink deep brown deep violet | 0.11 0.13 0.19 0.23 0.70 | deep black pink deep brown weak blue grey | 0.13 0.34 0.75 | orange violet weak brown | 0.20 0.28 0.34 0.67 0.75 | violet weak violet weak brown weak orange brown | 0.13 0.67 0.75 | weak blue gray brown |
| Sweet yellow clover (Melilotus officinalis) | 0.08 0.11 0.17 0.22 0.29 0.39 | deep violet deep brown yellow weak brown weak violet violet | 0.08 0.11 0.22 0.31 0.39 0.36 | deep violet deep purple weak brown pink orange weak green | 0.08 0.11 0.22 0.31 0.39 0.36 | deep pink deep purple brown pink orange blue | 0.05 0.15 0.37 | violet yellow violet | 0.05 0.14 0.32 | deep pink weak blue gray | 0.08 0.12 0.15 0.37 0.46 | weak pink weak brown weak gray orange pink |
| Clover (Trifolium L.) | 0.08 0.11 0.22 0.29 0.39 | deep violet deep brown weak brown weak violet violet | 0.08 0.11 0.22 0.31 0.39 | deep violet deep purple weak brown pink orange | 0.08 0.11 0.22 0.31 0.39 | deep pink deep purple brown pink orange | 0.06 0.11 0.30 | deep violet gray violet | 0.06 0.11 0.23 0.36 | weak blue pink weak orange weak orange | 0.06 0.11 0.13 0.26 0.29 | pink deep pink weak blue weak pink pink |
| Dandelion honey (Taraxacum spp.) | 0.15 0.40 0.36 | yellow deep yellow violet | 0.10 0.22 0.33 0.38 0.48 | deep purple brown brown deep pink weak gray | 0.10 0.16 0.21 0.38 | deep purple blue brown deep pink | 0.10 0.17 0.22 0.37 | green deep yellow weak green deep pink | 0.15 0.31 0.37 0.40 | blue weak brown deep pink gray | 0.10 0.16 0.37 | gray blue deep pink |
| Milk thistle (Silybum marianum (L.) Gaertner) | 0.11 0.22 0.29 0.36 0.39 | deep green weak brown weak violet orange violet | 0.11 0.22 0.36 0.49 | deep purple weak brown orange weak brown | 0.11 0.28 0.22 0.49 0.36 | deep purple weak pink brown gray orange | 0.13 0.28 0.32 0.36 | weak gray weak violet violet orange | 0.11 0.25 0.28 0.36 0.48 | deep pink weak gray gray orange weak brown | 0.07 0.14 0.25 0.36 | deep pink weak blue weak gray orange |
| Rape (Brassica napus L.) | 0.11 0.21 0.34 | deep green weak blue weak violet | 0.11 0.21 0.31 0.39 0.49 | deep brown gray deep gray pink gray | 0.11 0.22 0.31 0.40 0.50 | brown weak brown gray pink gray | 0.36 0.49 | weak green weak orange | 0.09 0.39 0.49 | weak orange pink gray | 0.09 0.21 0.31 0.49 | weak orange weak pink green gray |
| Samples | Declared Botanical Origin | Predominant Pollen (%) a | Botanical Origin | |
|---|---|---|---|---|
| R-1 | Rape | 89 6 | Brassica napus L. Salix spp. | Rape honey |
| R-2 | Rape | 73 18 | Brassica napus L. Salix spp. | Rape honey |
| W-1 | Willow | 61 27 | Brassica napus L. Salix spp. | Rape honey |
| W-2 | Willow | 59 20 | Salix spp. Brassica napus L. | Willow honey |
| W-3 | Willow | 65 10 | Salix spp. Rubus spp. | Willow honey * |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Makowicz, E.; Jasicka-Misiak, I.; Teper, D.; Kafarski, P. HPTLC Fingerprinting—Rapid Method for the Differentiation of Honeys of Different Botanical Origin Based on the Composition of the Lipophilic Fractions. Molecules 2018, 23, 1811. https://doi.org/10.3390/molecules23071811
Makowicz E, Jasicka-Misiak I, Teper D, Kafarski P. HPTLC Fingerprinting—Rapid Method for the Differentiation of Honeys of Different Botanical Origin Based on the Composition of the Lipophilic Fractions. Molecules. 2018; 23(7):1811. https://doi.org/10.3390/molecules23071811
Chicago/Turabian StyleMakowicz, Ewa, Izabela Jasicka-Misiak, Dariusz Teper, and Paweł Kafarski. 2018. "HPTLC Fingerprinting—Rapid Method for the Differentiation of Honeys of Different Botanical Origin Based on the Composition of the Lipophilic Fractions" Molecules 23, no. 7: 1811. https://doi.org/10.3390/molecules23071811
APA StyleMakowicz, E., Jasicka-Misiak, I., Teper, D., & Kafarski, P. (2018). HPTLC Fingerprinting—Rapid Method for the Differentiation of Honeys of Different Botanical Origin Based on the Composition of the Lipophilic Fractions. Molecules, 23(7), 1811. https://doi.org/10.3390/molecules23071811