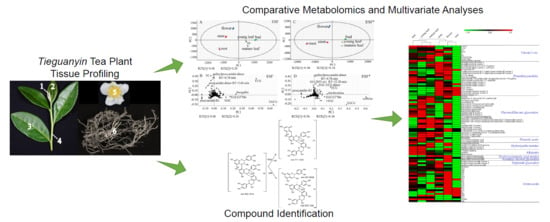
Insights into Tissue-specific Specialized Metabolism in Tieguanyin Tea Cultivar by Untargeted Metabolomics
Abstract
:1. Introduction
2. Results
2.1. Prominent Metabolite Variations Observed between Tea Plant Tissues
2.2. Structural Compositions of Oligomeric Proanthocyanidins Varied by Tissue Types
2.3. Flavonol Glycosides with Different Aglycone Moieties Displayed Spatial Distribution
2.4. Distribution of Purine Alkaloids and Hydroxycinnamic Acid Amides, Two Classes of Nitrogenous Compounds, Displayed Tissue Specificity
2.5. Differential Amino Acid Profiles among Tea Plant Tissues
3. Discussion
3.1. The Abundance of Flavonol Glycosides Demonstrates Tissue-specific Variations in Different Plants
3.2. Coumaroyl-Conjugated Hydroxycinnamic Acid Amides (HCCAs) are Unique Flower Metabolites
3.3. Occurrence of A-Type PAs is Rare in Tea Plants but Warrants Further Analysis
4. Materials and Methods
4.1. Plant Materials and Sampling
4.2. Extraction and UPLC-QTOF MS Analysis
4.3. Amino Acid Quantitation by UPLC-QqQ MS
4.4. Data Processing, Metabolite Identification, and Statistical Analysis
4.5. Chemicals and Reagents
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hayat, K.; Iqbal, H.; Malik, U.; Bilal, U.; Mushtaq, S. Tea and its consumption: Benefits and risks. Crit. Rev. Food Sci. Nutr. 2015, 55, 939–954. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.W.; Ogita, S.; Ashihara, H. Ethylamine content and theanine biosynthesis in different organs of Camellia sinensis seedlings. Naturforsch. C 2009, 64, 387–390. [Google Scholar] [CrossRef]
- Ashihara, H.; Deng, W.W.; Mullen, W.; Crozier, A. Distribution and biosynthesis of flavan-3-ols in Camellia sinensis seedlings and expression of genes encoding biosynthetic enzymes. Phytochemistry 2010, 71, 559–566. [Google Scholar] [CrossRef] [PubMed]
- Wu, Z.J.; Li, X.H.; Liu, Z.W.; Xu, Z.S.; Zhuang, J. De novo assembly and transcriptome characterization: Novel insights into catechins biosynthesis in Camellia sinensis. BMC Plant Biol. 2014, 14, 277. [Google Scholar] [CrossRef] [PubMed]
- Deng, W.W.; Ashihara, H. Occurrence and de novo biosynthesis of caffeine and theanine in seedlings of tea (Camellia sinensis). Nat. Prod. Commun. 2015, 10, 703–706. [Google Scholar] [PubMed]
- Liu, Z.W.; Wu, Z.J.; Li, H.; Wang, Y.X.; Zhuang, J. l-Theanine content and related gene expression: Novel insights into theanine biosynthesis and hydrolysis among different tea plant (Camellia sinensis L.) tissues and cultivars. Front. Plant. Sci. 2017, 8. [Google Scholar] [CrossRef] [PubMed]
- Wei, C.; Yang, H.; Wang, S.; Zhao, J.; Liu, C.; Gao, L.; Xia, E.; Lu, Y.; Tai, Y.; She, G.; et al. Draft genome sequence of Camellia sinensis var. sinensis provides insights into the evolution of the tea genome and tea quality. Proc. Natl. Acad. Sci. 2018, 20. [Google Scholar] [CrossRef]
- Wang, W.; Zhou, Y.; Wu, Y.; Dai, X.; Liu, Y.; Qian, Y.; Li, M.; Jiang, X.; Wang, Y.; Gao, L.; et al. Insight into catechins metabolic pathways of Camellia sinensis based on genome and transcriptome analysis. J. Agric. Food Chem. 2018, 1. [Google Scholar] [CrossRef] [PubMed]
- Xia, E.H.; Zhang, H.B.; Sheng, J.; Li, K.; Zhang, Q.J.; Kim, C.; Zhang, Y.; Liu, Y.; Zhu, T.; Li, W.; et al. The tea tree genome provides insights into tea flavor and independent evolution of caffeine biosynthesis. Mol. Plant 2017, 10, 866–877. [Google Scholar] [CrossRef] [PubMed]
- Wink, M. Evolution of secondary metabolites from an ecological and molecular phylogenetic perspective. Phytochemistry 2003, 64, 3–19. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, Y.; Li, W.; Zhao, L.; Meng, F.; Wang, Y.; Tan, H.; Yang, H.; Wei, C.; Wan, X.; et al. Tissue-specific, development-dependent phenolic compounds accumulation profile and gene expression pattern in tea plant Camellia sinensis. PLoS ONE 2013, 8, e62315. [Google Scholar] [CrossRef] [PubMed]
- Li, C.F.; Zhu, Y.; Yu, Y.; Zhao, Q.Y.; Wang, S.J.; Wang, X.C.; Yao, M.Z.; Luo, D.; Li, X.; Chen, L.; et al. Global transcriptome and gene regulation network for secondary metabolite biosynthesis of tea plant (Camellia sinensis). BMC Genom. 2015, 16, 560. [Google Scholar] [CrossRef] [PubMed]
- Suzuki, T.; Takahashi, E. Biosynthesis of caffeine by tea-leaf extracts. Enzymic formation of theobromine from 7-methylxanthine and of caffeine from theobromine. Biochem. J. 1975, 146, 87–96. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Deng, W.-W.; Ogita, S.; Ashihara, H. Biosynthesis of theanine (γ-ethylamino-L-glutamic acid) in seedlings of Camellia sinensis. Phytochem. Lett. 2008, 1, 115–119. [Google Scholar] [CrossRef]
- Deng, W.W.; Ashihara, H. Profiles of purine metabolism in leaves and roots of Camellia sinensis seedlings. Plant Cell Physiol. 2010, 51, 2105–2118. [Google Scholar] [CrossRef] [PubMed]
- Hanhineva, K.; Rogachev, I.; Kokko, H.; Mintz-Oron, S.; Venger, I.; Karenlampi, S.; Aharoni, A. Non-targeted analysis of spatial metabolite composition in strawberry (Fragariaxananassa) flowers. Phytochemistry 2008, 69, 2463–2481. [Google Scholar] [CrossRef] [PubMed]
- Asiago, V.M.; Hazebroek, J.; Harp, T.; Zhong, C. Effects of genetics and environment on the metabolome of commercial maize hybrids: A multisite study. J. Agric. Food Chem. 2012, 60, 11498–11508. [Google Scholar] [CrossRef] [PubMed]
- Jang, Y.K.; Jung, E.S.; Lee, H.-A.; Choi, D.; Lee, C.H. Metabolomic characterization of hot pepper (Capsicum annuum “CM334”) during fruit development. J. Agric. Food Chem. 2015, 63, 9452–9460. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Tohge, T.; Cuadros-Inostroza, A.; Tong, H.; Tenenboim, H.; Kooke, R.; Meret, M.; Keurentjes, J.B.; Nikoloski, Z.; Fernie, A.R.; et al. Mapping the Arabidopsis metabolic landscape by untargeted metabolomics at different environmental conditions. Mol. Plant 2018, 11, 118–134. [Google Scholar] [CrossRef] [PubMed]
- Liu, T. Chinese Tea; China Intercontinental Press: Beijing, China, 2005. [Google Scholar]
- Chen, Y.J.; Kuo, P.C.; Yang, M.L.; Li, F.Y.; Tzen, J.T.C. Effects of baking and aging on the changes of phenolic and volatile compounds in the preparation of old Tieguanyin oolong teas. Food Res. Int. 2013, 53, 732–743. [Google Scholar] [CrossRef]
- Meng, W.; Xu, X.; Cheng, K.-K.; Xu, J.; Shen, G.; Wu, Z.; Dong, J. Geographical origin discrimination of oolong Tea (TieGuanYin, Camellia sinensis (L.) O. Kuntze) using proton nuclear magnetic resonance spectroscopy and near-infrared spectroscopy. Food. Anal. Method 2017, 10, 3508–3522. [Google Scholar] [CrossRef]
- Li, Y.; Lei, J.; Yang, J.; Liu, R. Classification of Tieguanyin tea with an electronic tongue and pattern recognition. Anal. Lett. 2014, 47, 2361–2369. [Google Scholar] [CrossRef]
- Yan, S.M.; Liu, J.P.; Xu, L.; Fu, X.S.; Cui, H.F.; Yun, Z.Y.; Yu, X.P.; Ye, Z.H. Rapid discrimination of the geographical origins of an oolong tea (anxi-tieguanyin) by near-infrared spectroscopy and partial least squares discriminant analysis. J. Anal. Methods. Chem. 2014, 2014. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.Q.; Liu, P.P.; Shi, J.; Gao, Y.; Wang, Q.S.; Yin, J.F. Quality development and main chemical components of Tieguanyin oolong teas processed from different parts of fresh shoots. Food Chem. 2018, 249, 176–183. [Google Scholar] [CrossRef] [PubMed]
- Dou, J.; Lee, V.S.; Tzen, J.T.; Lee, M.R. Identification and comparison of phenolic compounds in the preparation of oolong tea manufactured by semifermentation and drying processes. J. Agric. Food Chem. 2007, 55, 7462–7468. [Google Scholar] [CrossRef] [PubMed]
- Tautenhahn, R.; Cho, K.; Uritboonthai, W.; Zhu, Z.; Patti, G.J.; Siuzdak, G. An accelerated workflow for untargeted metabolomics using the METLIN database. Nat. Biotechnol. 2012, 30, 826–828. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dai, W.D.; Qi, D.D.; Yang, T.; Lv, H.P.; Guo, L.; Zhang, Y.; Zhu, Y.; Peng, Q.H.; Xie, D.C.; Tan, J.F.; et al. Nontargeted analysis using ultraperformance liquid chromatography-quadrupole time-of-flight mass spectrometry uncovers the effects of harvest season on the metabolites and taste quality of tea (Camellia sinensis L.). J. Agric. Food Chem. 2015, 63, 9869–9878. [Google Scholar] [CrossRef] [PubMed]
- Fraser, K.; Harrison, S.J.; Lane, G.A.; Otter, D.E.; Hemar, Y.; Quek, S.-Y.; Rasmussen, S. HPLC–MS/MS profiling of proanthocyanidins in teas: A comparative study. J. Food Compos. Anal. 2012, 26, 43–51. [Google Scholar] [CrossRef]
- Jiang, X.; Liu, Y.; Wu, Y.; Tan, H.; Meng, F.; Wang, Y.S.; Li, M.; Zhao, L.; Liu, L.; Qian, Y.; et al. Analysis of accumulation patterns and preliminary study on the condensation mechanism of proanthocyanidins in the tea plant [Camellia sinensis]. Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Yan, T.; Hu, G.S.; Wang, A.H.; Hong, Y.; Jia, J.M. Characterisation of proanthocyanidins from Schisandra chinensis seed coats by UPLC-QTOF/MS. Nat. Prod. Res. 2014, 28, 1834–1842. [Google Scholar] [CrossRef] [PubMed]
- Lin, G.M.; Lin, H.Y.; Hsu, C.Y.; Chang, S.T. Structural characterization and bioactivity of proanthocyanidins from indigenous cinnamon (Cinnamomum osmophloeum). J. Sci. Food Agric. 2016, 96, 4749–4759. [Google Scholar] [CrossRef] [PubMed]
- Kamiya, K.; Watanabe, C.; Endang, H.; Umar, M.; Satake, T. Studies on the constituents of bark of Parameria laevigata Moldenke. Chem. Pharm. Bull. 2001, 49, 551–557. [Google Scholar] [CrossRef] [PubMed]
- Sawada, Y.; Nakabayashi, R.; Yamada, Y.; Suzuki, M.; Sato, M.; Sakata, A.; Akiyama, K.; Sakurai, T.; Matsuda, F.; Aoki, T.; et al. RIKEN tandem mass spectral database (ReSpect) for phytochemicals: A plant-specific MS/MS-based data resource and database. Phytochemistry 2012, 82, 38–45. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ferreres, F.; Silva, B.M.; Andrade, P.B.; Seabra, R.M.; Ferreira, M.A. Approach to the study of C-glycosyl flavones by ion trap HPLC-PAD-ESI/MS/MS: Application to seeds of quince (Cydonia oblonga). Phytochem. Anal. 2003, 14, 352–359. [Google Scholar] [CrossRef] [PubMed]
- Zheng, Y.; Hu, X.; Zhai, Y.; Liu, J.; Wu, G.; Wu, L.; Shen, T.J. Pharmacokinetics and tissue distribution study of camellianin A and its major metabolite in rats by liquid chromatography with tandem mass spectrometry. J. Chromatogr. B 2015, 997, 200–209. [Google Scholar] [CrossRef] [PubMed]
- Xie, C.; Xu, L.Z.; Luo, X.Z.; Zhong, Z.; Yang, S.L. Flavonol glycosides from Lysimachia capillipes. J. Asian Nat. Prod. Res. 2002, 4, 17–23. [Google Scholar] [CrossRef] [PubMed]
- Kashiwada, Y.; Nonaka, G.-I.; Nishioka, I.; Yamagishi, T. Galloyl and hydroxycinnamoylglucoses from rhubarb. Phytochemistry 1988, 27, 1473–1477. [Google Scholar] [CrossRef]
- Horai, H.; Arita, M.; Kanaya, S.; Nihei, Y.; Ikeda, T.; Suwa, K.; Ojima, Y.; Tanaka, K.; Tanaka, S.; Aoshima, K.; et al. MassBank: A public repository for sharing mass spectral data for life sciences. J. Mass Spectrom. 2010, 45, 703–714. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Dong, F.; Baldermann, S.; Murata, A.; Tu, Y.; Asai, T.; Watanabe, N. Isolation and identification of spermidine derivatives in tea (Camellia sinensis) flowers and their distribution in floral organs. J. Sci. Food Agric. 2012, 92, 2128–2132. [Google Scholar] [CrossRef] [PubMed]
- Li, H.J.; Deinzer, M.L. Tandem mass spectrometry for sequencing proanthocyanidins. Anal. Chem. 2007, 79, 1739–1748. [Google Scholar] [CrossRef] [PubMed]
- Saito, K.; Yonekura-Sakakibara, K.; Nakabayashi, R.; Higashi, Y.; Yamazaki, M.; Tohge, T.; Fernie, A.R. The flavonoid biosynthetic pathway in Arabidopsis: Structural and genetic diversity. Plant. Physiol. Biochem. 2013, 72, 21–34. [Google Scholar] [CrossRef] [PubMed]
- Dai, W.; Tan, J.; Lu, M.; Xie, D.; Li, P.; Lv, H.; Zhu, Y.; Guo, L.; Zhang, Y.; Peng, Q.; et al. Nontargeted modification-specific metabolomics investigation of glycosylated secondary metabolites in tea (Camellia sinensis L.) based on liquid chromatography-high-resolution mass spectrometry. J. Agric. Food Chem. 2016, 64, 6783–6790. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.X.; Yang, W.J.; Ahammed, G.J.; Shen, C.; Yan, P.; Li, X.; Han, W.Y. Developmental changes in carbon and nitrogen metabolism affect tea quality in different leaf position. Plant Physiol. Biochem. 2016, 106, 327–335. [Google Scholar] [CrossRef] [PubMed]
- Matsuda, F.; Yonekura-Sakakibara, K.; Niida, R.; Kuromori, T.; Shinozaki, K.; Saito, K. MS/MS spectral tag-based annotation of non-targeted profile of plant secondary metabolites. Plant J. 2009, 57, 555–577. [Google Scholar] [CrossRef] [PubMed]
- Youhnovski, N.; Werner, C.; Hesse, M. N,N’,N”-Triferuloylspermidine, a new UV absorbing polyamine derivative from pollen of Hippeastrum x hortorum. Naturforsch. C 2001, 56, 526–530. [Google Scholar] [CrossRef]
- Michael, A.J. Biosynthesis of polyamines and polyamine-containing molecules. Biochem. J. 2016, 473, 2315–2329. [Google Scholar] [CrossRef] [PubMed]
- Afendi, F.M.; Okada, T.; Yamazaki, M.; Hirai-Morita, A.; Nakamura, Y.; Nakamura, K.; Ikeda, S.; Takahashi, H.; Altaf-Ul-Amin, M.; Darusman, L.K.; et al. KNApSAcK family databases: Integrated metabolite-plant species databases for multifaceted plant research. Plant Cell Physiol. 2012, 53. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics--the link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef] [PubMed]
- Dixon, R.A.; Strack, D. Phytochemistry meets genome analysis, and beyond. Phytochemistry 2003, 62, 815–816. [Google Scholar] [CrossRef]
- Engelhardt, U.H. Chemistry of Tea. In Reference Module in Chemistry, Molecular Sciences and Chemical Engineering; Elsevier: Braunschweig, Germany, 2013. [Google Scholar]
- Tohge, T.; de Souza, L.P.; Fernie, A.R. Current understanding of the pathways of flavonoid biosynthesis in model and crop plants. J. Exp. Bot. 2017, 68, 4013–4028. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Tohge, T.; Niida, R.; Saito, K. Identification of a flavonol 7-O-rhamnosyltransferase gene determining flavonoid pattern in Arabidopsis by transcriptome coexpression analysis and reverse genetics. J. Biol. Chem. 2007, 282, 14932–14941. [Google Scholar] [CrossRef] [PubMed]
- Yonekura-Sakakibara, K.; Tohge, T.; Matsuda, F.; Nakabayashi, R.; Takayama, H.; Niida, R.; Watanabe-Takahashi, A.; Inoue, E.; Saito, K. Comprehensive flavonol profiling and transcriptome coexpression analysis leading to decoding gene–metabolite correlations in Arabidopsis. Plant Cell 2008, 20, 2160–2176. [Google Scholar] [CrossRef] [PubMed]
- Routaboul, J.M.; Kerhoas, L.; Debeaujon, I.; Pourcel, L.; Caboche, M.; Einhorn, J.; Lepiniec, L. Flavonoid diversity and biosynthesis in seed of Arabidopsis thaliana. Planta 2006, 224, 96–107. [Google Scholar] [CrossRef] [PubMed]
- Lai, J.P.; Lim, Y.H.; Su, J.; Shen, H.M.; Ong, C.N. Identification and characterization of major flavonoids and caffeoylquinic acids in three Compositae plants by LC/DAD-APCI/MS. J. Chromatogr. B 2007, 848, 215–225. [Google Scholar] [CrossRef] [PubMed]
- Schmid, M.; Davison, T.S.; Henz, S.R.; Pape, U.J.; Demar, M.; Vingron, M.; Scholkopf, B.; Weigel, D.; Lohmann, J.U. A gene expression map of Arabidopsis thaliana development. Nat. Genet. 2005, 37, 501–506. [Google Scholar] [CrossRef] [PubMed]
- Ishihara, H.; Tohge, T.; Viehoever, P.; Fernie, A.R.; Weisshaar, B.; Stracke, R. Natural variation in flavonol accumulation in Arabidopsis is determined by the flavonol glucosyltransferase BGLU6. J. Exp. Bot. 2016, 67, 1505–1517. [Google Scholar] [CrossRef] [PubMed]
- Scharbert, S.; Hofmann, T. Molecular definition of black tea taste by means of quantitative studies, taste reconstitution, and omission experiments. J. Agric. Food Chem. 2005, 53, 5377–5384. [Google Scholar] [CrossRef] [PubMed]
- Macoy, D.M.; Kim, W.Y.; Lee, S.Y.; Kim, M.G. Biosynthesis, physiology, and functions of hydroxycinnamic acid amides in plants. Plant Biotechnol. Rep. 2015, 9, 269–278. [Google Scholar] [CrossRef]
- Facchini, P.J.; Hagel, J.; Zulak, K.G. Hydroxycinnamic acid amide metabolism: Physiology and biochemistry. Can. J. Bot. 2002, 80, 577–589. [Google Scholar] [CrossRef]
- Negri, G.; Teixeira, E.W.; Florêncio Alves, M.L.T.M.; Moreti, A.C.d.C.C.; Otsuk, I.P.; Borguini, R.G.; Salatino, A. Hydroxycinnamic acid amide derivatives, phenolic compounds and antioxidant activities of extracts of pollen samples from Southeast Brazil. J. Agric. Food Chem. 2011, 59, 5516–5522. [Google Scholar] [CrossRef] [PubMed]
- Pang, Y.Z.; Abeysinghe, I.S.B.; He, J.; He, X.Z.; Huhman, D.; Mewan, K.M.; Sumner, L.W.; Yun, J.F.; Dixon, R.A. Functional characterization of proanthocyanidin pathway enzymes from tea and their application for metabolic engineering. Plant Physiol. 2013, 161, 1103–1116. [Google Scholar] [CrossRef] [PubMed]
- Gu, L.; Kelm, M.A.; Hammerstone, J.F.; Beecher, G.; Holden, J.; Haytowitz, D.; Prior, R.L. Screening of foods containing proanthocyanidins and their structural characterization using LC-MS/MS and thiolytic degradation. J. Agric. Food Chem. 2003, 51, 7513–7521. [Google Scholar] [CrossRef] [PubMed]
- Hashimoto, F.; Nonaka, G.; Nishioka, I. Tannins and related compounds. XC. 8-C-Ascorbyl (-)-epigallocatechin 3-O-gallate and novel dimeric flavan-3-ols, oolonghomobisflavans A and B, from oolong tea. Chem. Pharm. Bull. 1989, 37, 3255–3263. [Google Scholar] [CrossRef]
- Kumar, N.S.; Bandara, B.M.R.; Hettihewa, S.K. Isolation of a tetrameric A-type proanthocyanidin containing fraction from fresh tea (Camellia sinensis) leaves using high-speed counter-current chromatography. J. Liq. Chromatogr. Relat. Technol. 2015, 38, 1571–1575. [Google Scholar] [CrossRef]
- Chen, S.; Li, M.; Zheng, G.; Wang, T.; Lin, J.; Wang, S.; Wang, X.; Chao, Q.; Cao, S.; Yang, Z.; et al. Metabolite profiling of 14 Wuyi Rock tea cultivars using UPLC-QTOF MS and UPLC-QqQ MS combined with chemometrics. Molecules 2018, 23, 104. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds are not available from the authors. |





| Compound | Tentative Assignments | Rt (min) | Detected [M − H]− (m/z) | Theoretical [M − H]− (m/z) | Mass Error (ppm) | Formula | MS/MS Fragments | Ref. |
|---|---|---|---|---|---|---|---|---|
| Flavan-3-ols | ||||||||
| 1 | GC | 3.84 | 305.0670 | 305.0661 | 2.95 | C15H14O7 | 219.0664, 179.0353, 167.0351, 139.0399, 125.0245 | Authentic standard b |
| 2 | EGC | 4.93 | 305.0677 | 305.0661 | 5.24 | C15H14O7 | 219.0667, 179.0349, 167.0351, 139.0402, 125.0245 | Authentic standard b |
| 3 | C | 5.36 | 289.0719 | 289.0712 | 2.42 | C15H14O6 | 245.0822, 203.0714, 125.0246 | Authentic standard b |
| 4 | EC | 6.27 | 289.0721 | 289.0712 | 3.11 | C15H14O6 | 245.0819, 203.0714, 123.0451 | Authentic standard b |
| 5 | EGCG | 6.35 | 457.0797 | 457.0771 | 5.69 | C22H18O11 | 305.0669, 169.0160, 125.0247 | Authentic standard b |
| 6 | EGCG3”Me | 7.42 | 471.0934 | 471.0927 | 1.49 | C23H20O11 | 305.0674, 287.0568, 183.0304, 161.0251, 125.0247 | Authentic standard b |
| 7 | ECG | 7.86 | 441.0828 | 441.0822 | 1.36 | C22H18O10 | 331.0462, 289.0720, 245.0819, 169.0147, 125.0245 | Authentic standard b |
| 8 | ECG3”Me | 8.92 | 455.0960 | 455.0978 | −3.96 | C23H20O10 | 289.0721, 183.0302 | [26] |
| 9 | epiafzelechin 3-gallate | 8.97 | 425.0881 | 425.0873 | 1.88 | C22H18O9 | 273.0761, 169.0140, 151.0029, 137.0245, 125.0243 | [27] |
| Proanthocyanidins | ||||||||
| 10 | prodelphinidin B isomer 1 | 3.34 | 609.1246 | 609.1244 | 0.33 | C30H26O14 | 483.0947, 441.0827, 423.0717, 305.0667 | [26] |
| 11 | prodelphinidin B isomer 2 | 4.11 | 609.1249 | 609.1244 | 0.82 | C30H26O14 | 483.0932, 441.0822, 423.0716, 305.0668 | [26] |
| 12 | EC-GC dimer | 4.80 | 593.1300 | 593.1295 | 0.84 | C30H26O13 | 425.0857, 423.0707, 305.0670, 289.0717, 125.0245 | [27] |
| 13 | prodelphinidin B-2 (or 4) 3′-O-gallate | 5.11 | 761.1352 | 761.1354 | −0.26 | C37H30O18 | 609.1236, 591.1135, 577.1348, 423.0718 | [26] |
| 14 | procyanidin trimer (B type) isomer 1 | 5.25 | 865.1962 | 865.1980 | −2.08 | C45H38O18 | 695.1369, 577.1319, 451.1034, 287.0553 | [16] |
| 15 | procyanidin trimer (B type) isomer 2 | 5.52 | 865.1966 | 865.1980 | −1.62 | C45H38O18 | 695.1389, 575.1181, 451.0982, 287.0561 | [16] |
| 16 | procyanidin dimer (B type) isomer 1 | 5.68 | 577.1349 | 577.1346 | 0.52 | C30H26O12 | 451.1031, 425.0873, 407.0766, 289.0717, 125.0243 | [28] |
| 17 | procyanidin B2 | 5.78 | 577.1326 | 577.1346 | −3.47 | C30H26O12 | 451.1022, 425.0864, 407.0763, 289.0713, 125.0243 | Authentic standard b |
| 18 | procyanidin tetramer (B type) isomer 1 | 5.88 | 1153.2599 | 1153.2614 | −1.30 | C60H50O24 | 1027.2271, 865.1966, 576.1259, 575.1178, 287.0546 | [29] |
| 19 | procyanidin tetramer (B type) isomer 2 | 5.97 | 1153.2599 | 1153.2614 | −1.30 | C60H50O24 | 1027.2234, 577.1329, 575.1175, 287.0557 | [29] |
| 20 | procyanidin trimer (B type) isomer 3 | 5.99 | 865.1957 | 865.1980 | −2.66 | C45H38O18 | 739.1646, 713.1482, 695.1387, 577.1292, 451.1020, 423.0711, 407.0760, 287.0557 | [16] |
| 21 | EGC-ECG dimer | 6.04 | 745.1394 | 745.1405 | −1.48 | C37H30O17 | 593.1265, 423.0709, 407.0763, 169.0137 | [30] |
| 22 | (E)C-(4→8)-(E)C-(2→7, 4→8)-(E)C | 6.49 | 863.1814 | 863.1823 | −1.04 | C45H36O18 | 711.1324, 693.1232, 575.1180, 573.1035, 287.0559, 285.0392 | [31] |
| 23 | procyanidin tetramer isomer 3 (B type) | 6.72 | 1153.2589 | 1153.2614 | −2.17 | C60H50O24 | 865.1940,575.1193,287.0553 | [29] |
| 24 | 3-galloylprocyanidin B1/3′-galloylprocyanidin B2 isomer 1 | 6.78 | 729.1455 | 729.1456 | −0.14 | C37H30O16 | 603.1136, 441.0826, 407.0768, 289.0716, 125.0244 | [30] |
| 25 | parameritannin A-1 | 6.92 | 1153.2572 a | 1153.2614 a | −3.64 | C60H48O24 | 1001.2155, 866.2023, 579.1450, 577.1265 | [32,33] |
| 26 | epiafzelechin 3-O-gallate-(4β→6)-epigallocatechin 3-O-gallate | 7.50 | 883.1722 a | 883.1722 a | 0.00 | C44H34O20 | 409.0919, 271.0606, 153.0190 | [27] |
| 27 | epiafzelechin epicatechin 3,3′-digallate | 8.14 | 867.1774 a | 867.1773 a | 0.12 | C44H34O19 | 547.1236, 393.0977, 299.0561, 267.0661, 255.0660, 243.0659, 231.0663 | [27] |
| 28 | 3-galloylprocyanidin B1/3′-galloylprocyanidin B2 isomer 2 | 8.86 | 729.1455 | 729.1456 | −0.14 | C37H30O16 | 603.1140, 577.1115, 441.0829, 417.1560, 407.0777 | [30] |
| Flavonol/Flavone Glycosides | ||||||||
| 29 | isovitexin glucoside | 6.08 | 595.1653 a | 595.1663 a | −1.68 | C27H30O15 | 473.1142, 433.1129, 313.0711 | [34,35] |
| 30 | apigenin 6-C-glucoside 8-C-arabinoside | 6.91 | 563.1402 | 563.1401 | 0.18 | C26H28O14 | 545.1306, 503.1185, 473.1089, 443.1089, 383.0767, 353.0663 | [35] |
| 31 | myricetin 3-robinobioside (or 3-neohesperidoside) | 6.93 | 627.1556 a | 627.1561 a | −0.80 | C27H30O17 | 481.1028, 319.0457 | [27] |
| 32 | myricetin 3-galactoside | 7.02 | 479.0828 | 479.0826 | 0.42 | C21H20O13 | 317.0284, 316.0232, 271.0249 | [26] |
| 33 | myricetin 3′-glucoside | 7.11 | 479.0828 | 479.0826 | 0.42 | C21H20O13 | 317.0283, 316.0232, 271.0250 | [26] |
| 34 | quercetin 3-O-galactosyl rutinoside | 7.21 | 771.1981 | 771.1984 | −0.39 | C33H40O21 | 611.1627, 465.1064, 301.0348, 300.0270 | [27] |
| 35 | camellianin B | 7.68 | 577.1551 | 577.1557 | −1.04 | C27H30O14 | 433.1134, 313.0717, 269.0445 | [36] |
| 36 | rutin | 7.70 | 609.1450 | 609.1456 | −1.85 | C27H30O16 | 301.0343, 300.0280 | Authentic standard b |
| 37 | kaempferol 3-O-galactosyl rutinoside | 7.72 | 757.2177 a | 757.2191 a | 0.00 | C33H40O20 | 595.1652, 449.1080, 287.0557 | [26] |
| 38 | tricetin | 7.89 | 303.0506 a | 303.0505 a | 0.33 | C15H10O7 | 285.0410 | [27] |
| 39 | kaempferol 3-O-glucosyl rutinoside | 8.00 | 757.2187 a | 757.2191 a | −0.53 | C33H40O20 | 595.1661, 449.1079, 287.0563 | [26] |
| 40 | kaempferol 3-O-rutinoside | 8.43 | 595.1667 a | 595.1663 a | 0.67 | C27H30O15 | 503.0271, 449.1084, 287.0562 | [27] |
| 41 | kaempferol galactoside | 8.51 | 447.0928 | 447.0927 | 0.22 | C21H20O11 | 285.0376, 284.0328 | [26] |
| 42 | isorhamnetin glucoside | 8.65 | 477.1038 | 477.1033 | 1.12 | C22H22O12 | 357.1347, 315.0504, 314.0435, 300.0271, 299.0203 | [28] |
| 43 | kaempferol glucoside | 8.78 | 447.0930 | 447.0927 | 0.67 | C21H20O11 | 285.0393, 284.0333 | Authentic standard b |
| 44 | capilliposide I isomer 1 | 9.93 | 1063.2920 | 1063.2931 | −1.03 | C48H56O27 | 917.2346, 771.1968,753.1868, 615.1923,609.1423, 531.1428, 458.1134, 447.0933, 301.0351, 300.0273 | [37] |
| 45 | capilliposide II isomer 1 | 10.18 | 1049.3125 a | 1049.3138 a | −1.24 | C48H56O26 | 887.2597, 741.2037, 595.1495, 287.0557 | [37] |
| 46 | quercetin 3-(4”-(E)-p-coumaroylrobinobioside)-7-rhamnoside isomer 1 | 10.24 | 903.2554 a | 903.2559 a | −0.55 | C42H46O22 | 757.1984, 449.1078, 303.0508, 147.0448 | [27] |
| 47 | capilliposide I isomer 2 | 10.59 | 1065.3074 a | 1065.3087 a | −1.22 | C48H56O27 | 919.2526, 617.2090, 449.1088, 303.0505 | [37] |
| 48 | capilliposide II isomer 2 | 10.87 | 1049.3136 a | 1049.3138 a | −0.19 | C48H56O26 | 887.2601, 741.2042, 595.1545, 287.0559 | [37] |
| 49 | quercetin 3-(4′′-(E)-p-coumaroyl robinobioside)-7-rhamnoside isomer 2 | 10.91 | 903.2546 a | 903.2559 a | −1.44 | C42H46O22 | 757.1981, 449.1070, 303.0505, 147.0449 | [27] |
| 50 | isorhamnetin 3 (or 7)-(6′′-p-coumaroylglucoside) | 10.98 | 623.1400 | 623.1401 | −0.16 | C31H28O14 | 477.1068, 315.0508, 300.0271, 299.0200 | [27] |
| 51 | 2′′-O-trans-p-coumaroylastragalin | 11.68 | 593.1296 | 593.1295 | 0.17 | C30H26O13 | 447.0938, 285.0407, 284.0325 | [28] |
| Phenolic Acids | ||||||||
| 52 | theogallin | 2.90 | 343.0679 | 343.0665 | 4.08 | C14H16O10 | 191.0564 | Authentic standard b |
| 53 | 3-p-coumaroylquinic acid | 5.18 | 337.0931 | 337.0923 | 2.37 | C16H18O8 | 163.0403 | [26] |
| 54 | 5-p-coumaroylquinic acid | 6.41 | 337.0930 | 337.0923 | 2.08 | C16H18O8 | 173.0459 | [26] |
| Hydrolysable Tannins | ||||||||
| 55 | monogalloyl glucose | 2.45 | 331.0672 | 331.0665 | 2.11 | C13H16O10 | 271.0461, 211.0248, 169.0144, 151.0040, 125.0244 | [16] |
| 56 | methyl 6-O-galloyl-β-d-glucose | 3.66 | 345.0827 | 345.0822 | 1.45 | C14H18O10 | 225.0406, 183.0299 | [27] |
| 57 | digalloyl glucose isomer 1 | 4.76 | 483.0780 | 483.0775 | 1.04 | C20H20O14 | 313.0578, 169.0139 | [38] |
| 58 | digalloyl glucose isomer 2 | 5.01 | 483.0779 | 483.0775 | 0.83 | C20H20O14 | 313.0559, 169.0142 | [38] |
| Alkaloids | ||||||||
| 59 | 7-methylxanthine | 2.84 | 167.0570 a | 167.0569 a | 0.60 | C6H6N4O2 | 124.0514 | [39] |
| 60 | theobromine | 3.80 | 181.0729 a | 181.0726 a | 1.66 | C7H8N4O2 | 163.0622, 138.0674 | Authentic standard b |
| 61 | caffeine | 5.60 | 195.0885 a | 195.0882 a | 1.54 | C8H10N4O2 | 138.0673 | Authentic standard b |
| Hydroxycinnamic Acid Amides | ||||||||
| 62 | di-p-coumaroylputrescine | 10.33 | 381.1816 a | 381.1814 a | 0.53 | C22H24N2O4 | 235.1412, 218.1179, 147.0448, 119.0653, 91.0701 | [27] |
| 63 | tri-p-coumaroylspermidine | 12.08 | 584.2750 a | 584.2761 a | −1.88 | C34H37N36O | 438.2451, 420.2353, 292.2150, 275.1895, 205.1208, 204.1178, 147.0609, 119.0655, 91.0702 | [40] |
| Amino Acids | ||||||||
| 64 | theanine | 1.43 | 173.0935 | 173.0926 | 5.20 | C7H14N2O3 | 155.0830, 128.0354 | Authentic standard b |
| Aromatic Alcohol Glycosides | ||||||||
| 65 | phenylethyl primeveroside | 7.10 | 415.1599 | 415.1604 | −1.20 | C19H28O10 | 283.1177, 149.0448 | [27] |
| Terpenoid Glycosides | ||||||||
| 66 | linalool oxide primeveroside | 8.59 | 463.2166 | 463.2179 | −2.81 | C21H36O11 | 331.1761 | [27] |
| 67 | linalool primeveroside isomer 1 | 11.25 | 447.2234 | 447.2230 | 0.89 | C21H36O10 | 315.1805 | [27] |
| 68 | linalool primeveroside isomer 2 | 11.53 | 447.2233 | 447.2230 | 0.67 | C21H36O10 | 421.1703 | [27] |
| Amino Acids | Bud | Young Leaf | Mature Leaf | Stem | Flower | Root |
|---|---|---|---|---|---|---|
| Theanine | 8.36 ± 0.41 c | 10.41 ± 1.07 b | 10.58 ± 0.91 b | 33.36 ± 0.25 a | 6.32 ± 0.81 d | 1.41 ± 0.08 e |
| Aspartate | 8.27 ± 0.37 ab | 7.08 ± 0.55 b | 7.81 ± 0.25 ab | 9.71 ± 1.60 a | 7.69 ± 0.74 ab | ND |
| Glutamate | 1.88 ± 0.06 c | 2.55 ± 0.14 b | 3.01 ± 0.10 a | 2.55 ± 0.13 b | 1.74 ± 0.07 c | 0.26 ± 0.01 d |
| Glutamine | 0.99 ± 0.02 c | 0.58 ± 0.04 d | 0.35 ± 0.02 de | 6.15 ± 0.27 a | 2.77 ± 0.14 b | 0.03 ± 0.00 e |
| Serine | 0.96 ± 0.03 b | 0.75 ± 0.06 bc | 0.42 ± 0.03 cd | 0.46 ± 0.07 c | 2.79 ± 0.37 a | 0.03 ± 0.01 d |
| Arginine | 0.47 ± 0.02 c | 0.70 ± 0.05 b | 0.09 ± 0.01 d | 0.45 ± 0.01 c | 1.55 ± 0.10 a | 0.05 ± 0.00 d |
| Asparagine | 0.41 ± 0.01 b | 0.03 ± 0.00 d | 0.01 ± 0.00 d | 0.10 ± 0.02 c | 0.98 ± 0.04 a | 0.01 ± 0.01 d |
| Threonine | 0.09 ± 0.00 b | 0.03 ± 0.01 cd | 0.02 ± 0.00 d | 0.08 ± 0.01 bc | 0.34 ± 0.05 a | ND |
| Histidine | 0.08 ± 0.00 c | 0.03 ± 0.00 d | 0.01 ± 0.00 de | 0.10 ± 0.01 b | 0.34 ± 0.02 a | 0.01 ± 0.00 e |
| Tryptophan | 0.08 ± 0.00 b | 0.02 ± 0.00 e | ND | 0.05 ± 0.00 c | 0.26 ± 0.00 a | 0.03 ± 0.00 d |
| Valine | 0.07 ± 0.00 b | 0.06 ± 0.00 b | 0.03 ± 0.00 c | 0.06 ± 0.00 b | 0.35 ± 0.02 a | 0.01 ± 0.00 d |
| Lysine | 0.06 ± 0.00 c | 0.06 ± 0.00 c | 0.03 ± 0.00 d | 0.10 ± 0.01 b | 0.28 ± 0.01 a | 0.01 ± 0.00 e |
| Proline | 0.05 ± 0.00 b | 0.03 ± 0.00 b | 0.02 ± 0.00 b | 0.02 ± 0.00 b | 4.36 ± 0.06 a | ND |
| Leucine | 0.02 ± 0.00 b | 0.01 ± 0.00 b | 0.02 ± 0.00 b | 0.02 ± 0.00 b | 0.14 ± 0.01 a | ND |
| Isoleucine | 0.02 ± 0.00 b | 0.01 ± 0.00 bc | ND | 0.01 ± 0.00 bc | 0.36 ± 0.01 a | ND |
| Phenylalanine | 0.01 ± 0.00 c | 0.01 ± 0.00 c | 0.02 ± 0.00 c | 0.07 ± 0.00 b | 1.30 ± 0.03 a | ND |
| Methionine | 0.01 ± 0.00 b | ND | ND | 0.01 ± 0.00 b | 0.08 ± 0.01 a | ND |
| Tyrosine | ND | ND | ND | 0.01 ± 0.00 b | 0.13 ± 0.01 a | ND |
| γ-Aminobutyric acid | ND | ND | ND | 0.03 ± 0.00 c | 0.20 ± 0.01 a | 0.06 ± 0.00 b |
| Alanine | ND | 0.01 ± 0.01 b | ND | ND | 1.15 ± 0.25 a | ND |
| total | 21.82 ± 0.94 c | 22.38 ± 1.94 c | 22.43 ± 1.32 c | 53.35 ± 2.40 a | 33.14 ± 2.76 b | 1.92 ± 0.11 d |
© 2018 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chen, S.; Lin, J.; Liu, H.; Gong, Z.; Wang, X.; Li, M.; Aharoni, A.; Yang, Z.; Yu, X. Insights into Tissue-specific Specialized Metabolism in Tieguanyin Tea Cultivar by Untargeted Metabolomics. Molecules 2018, 23, 1817. https://doi.org/10.3390/molecules23071817
Chen S, Lin J, Liu H, Gong Z, Wang X, Li M, Aharoni A, Yang Z, Yu X. Insights into Tissue-specific Specialized Metabolism in Tieguanyin Tea Cultivar by Untargeted Metabolomics. Molecules. 2018; 23(7):1817. https://doi.org/10.3390/molecules23071817
Chicago/Turabian StyleChen, Si, Jun Lin, Huihui Liu, Zhihong Gong, Xiaxia Wang, Meihong Li, Asaph Aharoni, Zhenbiao Yang, and Xiaomin Yu. 2018. "Insights into Tissue-specific Specialized Metabolism in Tieguanyin Tea Cultivar by Untargeted Metabolomics" Molecules 23, no. 7: 1817. https://doi.org/10.3390/molecules23071817
APA StyleChen, S., Lin, J., Liu, H., Gong, Z., Wang, X., Li, M., Aharoni, A., Yang, Z., & Yu, X. (2018). Insights into Tissue-specific Specialized Metabolism in Tieguanyin Tea Cultivar by Untargeted Metabolomics. Molecules, 23(7), 1817. https://doi.org/10.3390/molecules23071817