Aldose Reductase, Protein Glycation Inhibitory and Antioxidant of Peruvian Medicinal Plants: The Case of Tanacetum parthenium L. and Its Constituents
Abstract
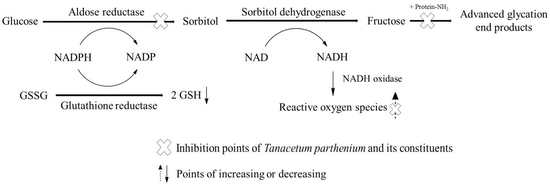
:1. Introduction
2. Results
2.1. Evaluation of Rat Lens Aldose Reductase and DPPH Radical Scavenging of 22 Peruvian Plant Extract
2.2. Ultrafiltration High-performance Liquid Chromatography Screening of AR Inhibitors in Tanacetum parthenium L
2.3. Screening Antioxidant Activity in Tanacetum parthenium L. by DPPH High-performance Liquid Chromatography Analysis
2.4. Structural Determination of Isolated Compounds
2.5. Inhibitory Effects of the Isolated Compounds on Rat Lens Aldose Reductase
2.6. Inhibitory Activities on the Sorbitol Accumulation by Active Compounds
2.7. DPPH Radical Scavenging Activity of the Isolated Compounds
2.8. Inhibitory Effects of the Isolated Compounds on Advanced Glycation End Products Formation
2.9. Interaction Analysis of Isolated Compounds with Aldose Reductase
3. Discussion
4. Materials and Methods
4.1. Chemicals and Reagents
4.2. Nuclear Magnetic Resonance and Mass Spectrometry Analysis
4.3. Plant Materials
4.4. Extraction and Isolation
4.5. High-performance Liquid Chromatography Analysis
4.6. Human Recombinant Aldose Reductase Ultrafiltration High-performance Liquid Chromatography Assay
4.7. Preparation of Rat Lens Aldose Reductase
4.8. Determination of Rat Lens Aldose Reductase Inhibition
4.9. Lens Culture and Intracellular Sorbitol Measurement
4.10. DPPH High-performance Liquid Chromatography Assay
4.11. Evaluation of DPPH Radical Scavenging Capacity
4.12. Bovine Serum Albumin–Methylglyoxal Assay for Advanced Glycation End Products Formation
4.13. Statistical Analysis
5. Conclusions
Author Contributions
Funding
Conflicts of Interest
Abbreviations
| AGEs | advanced glycation end products |
| AR | aldose reductase |
| ARIs | aldose reductase inhibitors |
| BSA | bovine serum albumin |
| COSY | correlation spectroscopy |
| DPPH | 2,2-diphenyl-1-picrylhydrazyl |
| DMSO | dimethylsulfoxide |
| HMBC | heteronuclear multiple bond correlation |
| HPLC | high-performance liquid chromatography |
| HRAR | human recombinant aldose reductase |
| HSQC | heteronuclear multiple quantum coherence |
| MeOH | methanol |
| NADPH | nicotinamide adenine dinucleotide phosphate |
| NMR | nuclear magnetic resonance |
| PAR | peak area reduction |
| RLAR | rat lens aldose reductase. |
| TBD | total binding degree |
| TP | Tanacetum parthenium |
References
- Petrash, J. All in the family: Aldose reductase and closely related aldo-keto reductases. Cell. Mol. Life Sci. Cmls 2004, 61, 737–749. [Google Scholar] [CrossRef] [PubMed]
- Akileshwari, C.; Raghu, G.; Muthenna, P.; Mueller, N.H.; Suryanaryana, P.; Petrash, J.M.; Reddy, G.B. Bioflavonoid ellagic acid inhibits aldose reductase: Implications for prevention of diabetic complications. J. Funct. Foods 2014, 6, 374–383. [Google Scholar] [CrossRef]
- Fowler, M.J. Microvascular and macrovascular complications of diabetes. Clin. Diabetes 2008, 26, 77–82. [Google Scholar] [CrossRef]
- Irani, M.; Minkoff, H.; Seifer, D.B.; Merhi, Z. Vitamin d increases serum levels of the soluble receptor for advanced glycation end products in women with pcos. J. Clin. Endocrinol. Metab. 2014, 99, E886–E890. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.H.; Wang, Z.; Lim, S.S. Chemo-enzymatic synthesis of vinyl and l-ascorbyl phenolates and their inhibitory effects on advanced glycation end products. Food Chem. 2017, 214, 726–735. [Google Scholar] [CrossRef]
- Ott, C.; Jacobs, K.; Haucke, E.; Navarrete Santos, A.; Grune, T.; Simm, A. Role of advanced glycation end products in cellular signaling. Redox Biol. 2014, 2, 411–429. [Google Scholar] [CrossRef] [Green Version]
- Robards, K. Strategies for the determination of bioactive phenols in plants, fruit and vegetables. J. Chromatography. A 2003, 1000, 657–691. [Google Scholar] [CrossRef]
- Dai, X.; Huang, Q.; Zhou, B.; Gong, Z.; Liu, Z.; Shi, S. Preparative isolation and purification of seven main antioxidants from eucommia ulmoides oliv. (du-zhong) leaves using hsccc guided by dpph-hplc experiment. Food Chem. 2013, 139, 563–570. [Google Scholar] [CrossRef]
- Xiao, S.; Yu, R.; Ai, N.; Fan, X. Rapid screening natural-origin lipase inhibitors from hypolipidemic decoctions by ultrafiltration combined with liquid chromatography-mass spectrometry. J. Pharm. Biomed. Anal. 2015, 104, 67–74. [Google Scholar] [CrossRef]
- Qiu, J.; Chen, X.; Netrusov, A.I.; Zhou, Q.; Guo, D.; Liu, X.; He, H.; Xin, X.; Wang, Y.; Chen, L. Screening and identifying antioxidative components in ginkgo biloba pollen by dpph-hplc-pad coupled with hplc-esi-ms2. Plos One 2017, 12, e0170141. [Google Scholar] [CrossRef]
- Li, S.; Li, S.; Tang, Y.; Liu, C.; Chen, L.; Zhang, Y. Ultrafiltration-lc–ms combined with semi-preparative hplc for the simultaneous screening and isolation of lactate dehydrogenase inhibitors from belamcanda chinensis. J. Sep. Sci. 2016, 39, 4533–4543. [Google Scholar] [CrossRef]
- Zhao, H.; Zhang, Y.; Guo, Y.; Shi, S. Identification of major α-glucosidase inhibitors in radix astragali and its human microsomal metabolites using ultrafiltration hplc–dad–msn. J. Pharm. Biomed. Anal. 2015, 104, 31–37. [Google Scholar] [CrossRef]
- de Almeida, L.M.S.; Carvalho, L.S.A.d.; Gazolla, M.C.; Silva Pinto, P.L.; Silva, M.P.N.d.; de Moraes, J.; Da Silva Filho, A.A. Flavonoids and sesquiterpene lactones from artemisia absinthium and tanacetum parthenium against schistosoma mansoni worms. Evid. Based Complement. Altern. Med. 2016, 2016. [Google Scholar] [CrossRef]
- Qing, W.; Wang, Y.; Li, H.; Zhu, J.; Liu, X. Hydrogels generated by low-molecular-weight pegylated luteolin and α-cyclodextrin through self-assembly for 5-fluorouracil delivery. Rsc Adv. 2016, 6, 95812–95817. [Google Scholar] [CrossRef]
- Kim, H.-J.; Jeong, H.-S.; Kang, S.-H. Taxonomical study of chrysosplenium l.(saxifragaceae) in korea based on chemical composition. Korean J. Plant. Resour. 2013, 26, 718–725. [Google Scholar] [CrossRef]
- Jang, Y.S.; Wang, Z.; Lee, J.-M.; Lee, J.-Y.; Lim, S.S. Screening of korean natural products for anti-adipogenesis properties and isolation of kaempferol-3-o-rutinoside as a potent anti-adipogenetic compound from solidago virgaurea. Molecules 2016, 21, 226. [Google Scholar] [CrossRef]
- Kim, J.K.; Lee, Y.S.; Kim, S.H.; Bae, Y.S.; Lim, S.S. Inhibition of aldose reductase by phenylethanoid glycoside isolated from the seeds of paulownia coreana. Biol. Pharm. Bull. 2011, 34, 160–163. [Google Scholar] [CrossRef]
- Yin, X.F.; Jeon, Y.E.; Chung, H.C.; Choung, S.Y.; Shim, J.-H.; Kang, I.-J. In vitro efficacy evaluation for prevention of diabetes and diabetic complications using aster sphathulifolius. Food Sci. Biotechnol. 2015, 24, 301–306. [Google Scholar] [CrossRef]
- Matsuda, H.; Wang, T.; Managi, H.; Yoshikawa, M. Structural requirements of flavonoids for inhibition of protein glycation and radical scavenging activities. Bioorganic Med. Chem. 2003, 11, 5317–5323. [Google Scholar] [CrossRef]
- Li, H.M.; Hwang, S.H.; Kang, B.G.; Hong, J.S.; Lim, S.S. Inhibitory effects of colocasia esculenta (l.) schott constituents on aldose reductase. Molecules 2014, 19, 13212–13224. [Google Scholar] [CrossRef]
- Chen, M.; Liu, L.; Chen, X. Preparative isolation and analysis of alcohol dehydrogenase inhibitors from glycyrrhiza uralensis root using ultrafiltration combined with high-performance liquid chromatography and high-speed countercurrent chromatography. J. Sep. Sci. 2014, 37, 1546–1551. [Google Scholar] [CrossRef]
- Prodanov, M.; Vacas, V.; Hernández, T.; Estrella, I.; Amador, B.; Winterhalter, P. Chemical characterisation of malvar grape seeds (vitis vinifera l.) by ultrafiltration and rp-hplc-pad-ms. J. Food Compos. Anal. 2013, 31, 284–292. [Google Scholar] [CrossRef]
- Liu, L.; Shi, S.; Chen, X.; Peng, M. Analysis of tyrosinase binders from glycyrrhiza uralensis root: Evaluation and comparison of tyrosinase immobilized magnetic fishing-hplc and reverse ultrafiltration-hplc. J. Chromatogr. B 2013, 932, 19–25. [Google Scholar] [CrossRef] [PubMed]
- Zhou, X.; Liang, J.; Zhang, Y.; Zhao, H.; Guo, Y.; Shi, S. Separation and purification of α-glucosidase inhibitors from polygonatum odoratum by stepwise high-speed counter-current chromatography combined with sephadex lh-20 chromatography target-guided by ultrafiltration–hplc screening. J. Chromatogr. B 2015, 985, 149–154. [Google Scholar] [CrossRef]
- Ma, R.; Zhou, R.; Tong, R.; Shi, S.; Chen, X. At-line hyphenation of high-speed countercurrent chromatography with sephadex lh-20 column chromatography for bioassay-guided separation of antioxidants from vine tea (ampelopsis grossedentata). J. Chromatogr. B 2017, 1040, 112–117. [Google Scholar] [CrossRef]
- Majdi, M.; Karimzadeh, G.; Malboobi, M. Spatial and developmental expression of key genes of terpene biosynthesis in tanacetum parthenium. Biol. Plant. 2014, 58, 379–384. [Google Scholar] [CrossRef]
- Végh, K.; Alberti, Á.; Riethmüller, E.; Tóth, A.; Béni, S.; Kéry, Á. Separation and analysis of bioactive flavonoids in tanacetum parthenium supercritical fluid extracts. Planta Med. 2015, 81, PM_56. [Google Scholar] [CrossRef]
- Orhan, I.E.; Tosun, F.; Gülpınar, A.R.; Kartal, M.; Duran, A.; Mihoglugil, F.; Akalgan, D. Lc–ms quantification of parthenolide and cholinesterase inhibitory potential of selected tanacetum l.(emend. Briq.) taxa. Phytochem. Lett. 2015, 11, 347–352. [Google Scholar] [CrossRef]
- Majdi, M.; Abdollahi, M.R.; Maroufi, A. Parthenolide accumulation and expression of genes related to parthenolide biosynthesis affected by exogenous application of methyl jasmonate and salicylic acid in tanacetum parthenium. Plant. Cell Rep. 2015, 34, 1909–1918. [Google Scholar] [CrossRef]
- Kim, S.B.; Hwang, S.H.; Suh, H.W.; Lim, S.S. Phytochemical analysis of agrimonia pilosa ledeb, its antioxidant activity and aldose reductase inhibitory potential. Int. J. Mol. Sci. 2017, 18, 379. [Google Scholar] [CrossRef]
- Hwang, S.H.; Kwon, S.H.; Kim, S.B.; Lim, S.S. Inhibitory activities of stauntonia hexaphylla leaf constituents on rat lens aldose reductase and formation of advanced glycation end products and antioxidant. BioMed Res. Int. 2017, 2017, 4273257. [Google Scholar] [CrossRef]
- Jung, H.A.; Islam, M.N.; Kwon, Y.S.; Jin, S.E.; Son, Y.K.; Park, J.J.; Sohn, H.S.; Choi, J.S. Extraction and identification of three major aldose reductase inhibitors from artemisia montana. Food Chem. Toxicol. 2011, 49, 376–384. [Google Scholar] [CrossRef] [PubMed]
- Jung, H.A.; Moon, H.E.; Oh, S.H.; Kim, B.-W.; Sohn, H.S.; Choi, J.S. Kinetics and molecular docking studies of kaempferol and its prenylated derivatives as aldose reductase inhibitors. Chem. -Biol. Interact. 2012, 197, 110–118. [Google Scholar] [CrossRef]
- Wang, Z.; Hwang, S.H.; Huang, B.; Lim, S.S. Identification of tyrosinase specific inhibitors from xanthium strumarium fruit extract using ultrafiltration-high-performance liquid chromatography. J. Chromatogr. B 2015, 1002, 319–328. [Google Scholar] [CrossRef]
- Li, H.M.; Kim, J.K.; Jang, J.M.; Cui, C.B.; Lim, S.S. Analysis of the inhibitory activity of abeliophyllum distichum leaf constituents against aldose reductase by using high-speed counter current chromatography. Arch. Pharm. Res. 2013, 36, 1104–1112. [Google Scholar] [CrossRef] [PubMed]
- Paek, J.H.; Shin, K.H.; Kang, Y.-H.; Lee, J.-Y.; Lim, S.S. Rapid identification of aldose reductase inhibitory compounds from perilla frutescens. BioMed Res. Int. 2013, 2013. [Google Scholar] [CrossRef]
- Lee, Y.S.; Kim, S.H.; Jung, S.H.; Kim, J.K.; Pan, C.-H.; Lim, S.S. Aldose reductase inhibitory compounds from glycyrrhiza uralensis. Biol. Pharm. Bull. 2010, 33, 917–921. [Google Scholar] [CrossRef]
- Lee, K.J.; Oh, Y.C.; Cho, W.K.; Ma, J.Y. Antioxidant and anti-inflammatory activity determination of one hundred kinds of pure chemical compounds using offline and online screening hplc assay. Evid. Based Complement. Altern. Med. 2015, 2015. [Google Scholar] [CrossRef]
- Li, H.M.; Kim, J.K.; Jang, J.M.; Kwon, S.O.; Cui, C.B.; Lim, S.S. The inhibitory effect of prunella vulgaris l. On aldose reductase and protein glycation. BioMed Res. Int. 2012, 2012. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |




| Catalogues | Family | Spices | Common Names | Used Part | Yield (%) | Inhibition (%) | |
|---|---|---|---|---|---|---|---|
| RLAR | DPPH | ||||||
| LNP-1 | PTERIDACEAE | Cheilanthes pruinata Kaulf. | Cuti-Cuti Marron macho | Aerial part | 30.3 | 41.6 ± 1.1 | 53.2 ± 0.3 |
| LNP-3 | FABACEAE | Otholobium mexicanum (L. f.) J.W. Grimes | Culen Negro | Aerial part | 6.2 | 50.4 ± 0.5 | 4.5 ± 5.7 |
| LNP-7 | FABACEAE | Senna sp. | Hoja de sen | Leaf | 8.2 | 29.6 ± 2.3 | NI a |
| LNP-11 | LAMIACEAE | Ocimum basilicum L. | Albahaca de olor | Leaf | 5.6 | 16.5 ± 0.8 | 6.0 ± 1.4 |
| LNP-13 | POACEAE | Chrysopogon zizanioides (L.) Roberty | Pachuli | Leaf | 9.1 | 5.8 ± 3.4 | NI |
| LNP-15 | LAMIACEAE | Clinopodium pulchellum (Kunth) Govaerts | Panisara | Leaf | 4.2 | 38.5 ± 2.5 | 52.8 ± 3.4 |
| LNP-18 | POACEAE | Cymbopogon citratus (DC.) Stapf | Hierba Luisa | Leaf | 6.9 | 3.2 ± 0.1 | 2.4 ± 1.3 |
| LNP-19 | SCROPHULAR-IACEAE | Buddleja americana L. | Flor Blanca | Flowers | 8.6 | 2.8± 0.2 | 2.8 ± 1.1 |
| LNP-20 | CARYOPHYL-LACEAE | Dianthus caryophyllus L. | Clavel | Leaf | 12.4 | 23.1 ± 3.4 | 6.4 ± 4.8 |
| LNP-23 | ASTERACEAE | Tanacetum parthenium (L.) Sch. Bip. | Santa Maria | Whole | 10.3 | 61.1 ± 0.5 | 88.6 ± 2.1 |
| LNP-24 | CAPRIFOLIAEAE | Sambucus peruviana H. B. K | Sauco (tilo) | Leaf | 8.9 | 51.3 ± 0.9 | 53.3 ± 1.6 |
| LNP-27 | LYCOPODIACE-AE | Huperzia crassa (Humb. & Bonpl. ex Willd.) Rothm. | Trensilla or enredadera | Leaf | 12.3 | 19.7 ± 0.9 | NI |
| LNP-28 | CYPERACEAE | Eleocharis albibracteata Nees & Meyen ex Kunth | Hierba del caballero | Leaf | 3.9 | 50.2 ± 1.4 | 23.6 ± 2.3 |
| LNP-33 | ASTERACEAE | Werneria nubigena Kunth | Condor | Leaf | 8.6 | 43.9 ± 1.8 | 11.4 ± 7.0 |
| LNP-39 | EUPHORBIACE-AE | Jatropha curcas L. | Pinones, pinol | Seed | 2.0 | 16.2 ± 3.2 | 4.3 ± 9.6 |
| LNP-41 | ANACARDIAC-EAE | Anacardium occidentale L. | Pepa de la selva, Casho | Fruit | 25.7 | 16.2 ± 0.2 | 36.7 ± 0.4 |
| LNP-43 | LAURACEAE | cf. Endlicheria | Spingo | Seed | 10.1 | 3.3 ± 0.4 | 2.3 ± 0.9 |
| LNP-45 | SOLANACEAE | Capsicum chinense | Aji panca rojo | Fruit | 42.0 | 2.1 ± 0.3 | 4.1 ± 0.3 |
| LNP-46 | SOLANACEAE | Capsicum baccatum | Aji amarillo | Fruit | 50.9 | 8.7 ± 0.7 | 1.1 ± 0.2 |
| LNP-47 | FABACEAE | Lupinus mutabilis | Tarwi | Seed | 10.2 | NI | NI |
| LNP-48 | URTICACEAE | Urtica magellanica A. Jussie ex Poiret | Ortiga Negra | Aerial part | 6.6 | NI | 4.3 ± 6.9 |
| LNP-80 | FABACEAE | Desmodium molliculum (Kunth) DC. | Manayupa | Leaf | 23.8 | 12.9 ± 0.5 | 90.0 ± 4.8 |
| Entry | Concentrations (μg/mL) | Inhibition (%) | IC50 (μg/mL) a | |
|---|---|---|---|---|
| RLAR | 70% MeOH | 10 | 61.10 | 8.04 ± 0.61 |
| 5 | 31.24 | |||
| 1 | 20.35 | |||
| Quercetin b | 10 | 81.28 | 5.35 ± 0.20 | |
| 1 | 44.80 | |||
| 0.5 | 23.56 | |||
| DPPH | 70% MeOH | 75 | 88.58 | 33.22 ± 2.09 |
| 30 | 49.53 | |||
| 15 | 31.84 | |||
| L-ascorbic acid c | 15 | 97.23 | 6.02 ± 0.37 | |
| 7.5 | 58.34 | |||
| 3 | 33.71 | |||
| AGEs | 70% MeOH | 200 | 61.01 | 163.71 ± 6.31 |
| 100 | 30.49 | |||
| 20 | 8.16 | |||
| Aminoguanidine d | 200 | 75.32 | 121.96 ± 5.10 | |
| 100 | 39.94 | |||
| 20 | 20.75 | |||
| Compounds | RLAR | DPPH | AGEs | ||
|---|---|---|---|---|---|
| IC50 (μM) a | TBD (%) b | IC50 (μM) | PAR (%) c | AGEs | |
| Ferulic acid (1) | 3.20 ±0.12 | 27.08 | 16.23 ± 0.41 | 23.53 | 5.59 ± 0.26 |
| Apigenin (2) | 1.97 ± 0.10 | 15.00 | 14.06 ± 0.72 | 11.28 | NI |
| Luteolin-7-O-glucoside (3) | 1.31 ± 0.09 | 26.58 | 6.44 ± 0.14 | 16.85 | 3.43 ± 0.12 |
| Luteolin (4) | 1.76 ± 0.03 | 38.02 | 11.84 ± 0.37 | 29.36 | 6.73 ± 0.43 |
| Chrysosplenol (5) | 1.92 ± 0.08 | 25.54 | >25 | 7.19 | NI |
| Kaempferol (6) | 1.11 ± 0.03 | 24.29 | 8.32 ± 0.54 | 13.97 | NI |
| Santin (7) | NI b | - | NI | - | NI |
| Quercetin d | 1.77 ± 0.53 | - | - | - | - |
| L-ascorbic acid e | - | - | 3.41 ± 0.11 | - | - |
| Aminoguanidine f | - | - | - | - | 110.55 ± 3.28 |
| Compounds | Sorbitol Content (mg)/lens Wet Weight (g) | Inhibition (%) |
|---|---|---|
| Sorbitol free | No detection | - |
| Control | 1.47 ± 0.04 | - |
| Quercetin a | 0.21 ± 0.02 | 85.71 ± 5.71 |
| Ferulic acid (1) | 0.29 ± 0.03 | 80.27 ± 2.38 |
| Apigenin (2) | 0.19 ± 0.06 | 87.07 ± 2.37 |
| Luteolin-7-O-glucoside (3) | 0.07 ± 0.01 | 95.23 ± 5.97 |
| Luteolin (4) | 0.12 ± 0.02 | 91.83 ± 5.23 |
| Chrysosplenol (5) | 0.26 ± 0.03 | 82.31 ± 2.39 |
| Kaempferol (6) | 0.03 ± 0.00 | 97.95 ± 6.31 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hwang, S.H.; Kim, H.-Y.; Guillen Quispe, Y.N.; Wang, Z.; Zuo, G.; Lim, S.S. Aldose Reductase, Protein Glycation Inhibitory and Antioxidant of Peruvian Medicinal Plants: The Case of Tanacetum parthenium L. and Its Constituents. Molecules 2019, 24, 2010. https://doi.org/10.3390/molecules24102010
Hwang SH, Kim H-Y, Guillen Quispe YN, Wang Z, Zuo G, Lim SS. Aldose Reductase, Protein Glycation Inhibitory and Antioxidant of Peruvian Medicinal Plants: The Case of Tanacetum parthenium L. and Its Constituents. Molecules. 2019; 24(10):2010. https://doi.org/10.3390/molecules24102010
Chicago/Turabian StyleHwang, Seung Hwan, Hyun-Yong Kim, Yanymee N. Guillen Quispe, Zhiqiang Wang, Guanglei Zuo, and Soon Sung Lim. 2019. "Aldose Reductase, Protein Glycation Inhibitory and Antioxidant of Peruvian Medicinal Plants: The Case of Tanacetum parthenium L. and Its Constituents" Molecules 24, no. 10: 2010. https://doi.org/10.3390/molecules24102010
APA StyleHwang, S. H., Kim, H. -Y., Guillen Quispe, Y. N., Wang, Z., Zuo, G., & Lim, S. S. (2019). Aldose Reductase, Protein Glycation Inhibitory and Antioxidant of Peruvian Medicinal Plants: The Case of Tanacetum parthenium L. and Its Constituents. Molecules, 24(10), 2010. https://doi.org/10.3390/molecules24102010