Four New Flavonoids Isolated from the Aerial Parts of Cadaba rotundifolia Forssk. (Qadab)
Abstract
:1. Introduction
2. Results and Discussion
2.1. Isolation and Spectroscopic Analyses of the Compounds
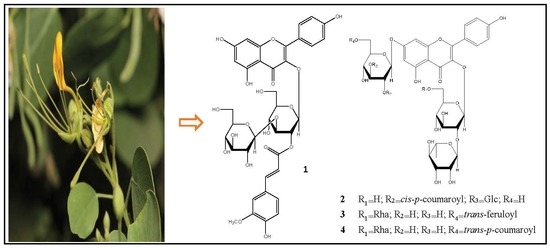
2.1.1. Chemical Structure of Compound 1
2.1.2. Chemical Structure of Compound 2
2.1.3. Chemical Structure of Compound 3
2.1.4. Chemical Structure of Compound 4
2.2. Biological Activities of the Isolated Compounds
3. Materials and Methods
3.1. General Methods
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Spectroscopic Data of Compounds 1–4
3.5. Acid Hydrolysis
3.6. Alkaline Hydrolysis
3.7. DPPH Radical Scavenging Activity
3.8. Determination of In Vitro AGEs Formation
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Hall, J.C.; Sytsma, K.J.; Iltis, H.H. Phylogeny of Capparaceae and Brassicaceae based on chloroplast sequence data. Am. J. Bot. 2002, 89, 1826–1842. Available online: https://doi.org/10.3732/ajb.89.11.1826 (accessed on 5 June 2019). [CrossRef] [PubMed]
- Kers, L.E. Capparceae. In The Families and Genera of Vascular Plants. Flowering Plants, Dicotyledons: Malvales, Capparales, and Non-betalain Caryophyllales.; Kubitzki, K., Bayer, C., Eds.; Springer: Berlin/Heidelberg, Germany, 2003; Volume 5, pp. 36–56. [Google Scholar]
- Migahid, A.; Hammouda, M. Flora of Saudi Arabia, 1st ed.; Riyadh University Publication: Riyadh, Saudi Arabia, 1974; p. 42. [Google Scholar]
- Ahmad, V.; Amber, A.; Arif, S.; Chen, M.H.; Clardy, J. Cadabicine, an alkaloid from Cadaba farinose. Phytochemistry 1985, 24, 2709–2711. Available online: https://doi.org/10.1016/S0031-9422(00)80700-1 (accessed on 5 June 2019). [CrossRef]
- Al-Musayeib, N.M.; Mohamed, G.A.; Ibrahim, S.R.; Ross, S.A. Lupeol-3-O-decanoate, a new triterpene ester from Cadaba farinose Frossk. Growing in Saudi Arabia. Med. Chem. Res. 2013, 22, 5297–5302. [Google Scholar] [CrossRef]
- Mohamed, G.A.; Ibrahim, S.R.; Al-Musayeib, N.M.; Ross, S.A. New anti-inflammatory flavonoids from Cadaba glandulosa Frossk. Arch. Pharm. Res. 2014, 37, 459–466. [Google Scholar] [CrossRef] [PubMed]
- Velmurugan, P.; Kamaraj, M.; Prema, D. Phytochemical constituents of Cadaba trifoliata Roxb. root extract. Int. J. Phytomed. 2010, 2, 379–384. [Google Scholar]
- Kamel, W.M.; El-Ghani, M.M.; El-Bous, M.M. Taxonomic study of Capparaceae from Egypt: Revisited. AJPSB 2009, 3, 27–35. [Google Scholar]
- Yousif, G.; Iskander, G.M.; Eisa, E. Alkaloid components in the Sudan flora. Part II. Alkaloid of Cadaba farinose and C. rotundifolia. Fitoterapia 1986, 55, 117–118. [Google Scholar]
- Harbaum, B.; Hubbermann, E.M.; Wolf, C.; Herges, R.; Zhu, Z.; Schwarz, K. Identification of flavonoids and hydroxycinnamic acid in pak choi varieties (Brassica campestris L. ssp. chinesis var. communis) by HPLC-ESI-MSn and NMR and their quantification by HPLC-DAD. J. Agric. Food Chem. 2007, 3, 8251–8260. [Google Scholar] [CrossRef] [PubMed]
- Gossan, D.P.; Alabdul Majed, A.; Yao-Kouassi, P.A.; Coffy, A.A.; Harakat, D.; Voutaquenne-Nazabadioko, L. New acylated flavonol glycosides from the aerial parts of Gouania longipetala. Phytochem. Lett. 2015, 11, 306–310. [Google Scholar] [CrossRef]
- Corea, G.; Fattorusso, E.; Lanzotti, V. Saponins and flavonoids of Allium triquetrum. J. Nat. Prod. 2003, 66, 1405–1411. [Google Scholar] [CrossRef] [PubMed]
- Carotenuto, A.; Feo, V.D.; Fattorusso, E.; Lanzotti, V.; Magnot, S.; Cicala, C. The flavonoids of Allium ursinum. Phytochemistry 1996, 41, 531–536. [Google Scholar] [CrossRef]
- Séro, L.; Sanguinet, L.; Blanchard, P.; Dang, B.T.; Morel, S.; Richomme, P.; Séraphin, D.; Séverine, D. Tuning a 96-well microtiter plate fluorescence-based assay to identify AGE inhibitors in crude plant extracts. Molecules 2013, 18, 14320–14339. [Google Scholar] [CrossRef] [PubMed]
- Sugimoto, S.; Wanas, A.S.; Mizuta, T.; Matsunami, K.; Kamel, M.S.; Otsuka, H. Structure elucidation of secondary metabolites isolated from the leaves of lxora undulate and their inhibitory activity toward advanced glycation end-products formation. Phytochemistry 2014, 108, 189–195. [Google Scholar] [CrossRef] [PubMed]
- Lee, K.T.; Choi, J.H.; Kim, D.H.; Son, K.H.; Kim, W.B.; Kown, S.H.; Park, H.J. Constituents and antitumor principle of Allium victorialis var. platyphyllum. Arch. Pharm. Res. 2011, 24, 44–50. [Google Scholar] [CrossRef]
- Kazuma, K.; Noda, N.; Suzuki, M. Malonylated flavonol glycosides from the petals of Clitoria ternatea. Phytochemistry 2003, 62, 229–237. [Google Scholar] [CrossRef]
- Walter, A.; Sequin, U. Flavonoids from the leaves of Boscia salicifolia. Phytochemistry 1990, 29, 2561–2563. [Google Scholar] [CrossRef]
- Mabry, T.J.; Markham, K.R.; Thomas, M.B. The Systematic Identification of Flavonoids; Springer: New York, NY, USA, 1970; pp. 253–273. [Google Scholar]
- Zheng, X.; Li, M.; Zeng, M.; Zhang, J.; Zhao, X.; Lv, J.; Zhang, Z.; Feng, W. Extraction Method of Beitingxinhuangtong C from Lipidium apetalum and Its Application in Preparing Estrogenic Drug. Faming Zhuanli Shenging Gongkai Shoumingshu CN 1076024640 A 201180119, 19 January 2018. [Google Scholar]
- Qin, X.; Xing, Y.F.; Zhou, Z.; Yao, Y. Dihydrochalcone compounds isolated from Crabapple leaves showed anticancer effects on human cancer cell line. Molecules 2015, 20, 21193–21230. [Google Scholar] [CrossRef] [PubMed]
- Yoshikawa, M.; Sugimoto, S.; Nakamura, S.; Matsuda, H. Medicinal flowers. XXII structures of chakasaponins V and VI, chakanoside I, and chakaflavonoside A from flower buds of Chinese tea plant (Camellia sinensis). Chem. Pharm. Bull. 2008, 56, 1297–1303. [Google Scholar] [CrossRef] [PubMed]
- Mitani, T.; Mimura, H.; Ikeda, K.; Nishide, M.; Yamaguchi, M.; Koyama, H.; Hayashi, Y.; Sakamoto, H. Process for the purification of cis-p-coumaric acid by cellulose column chromatography after the treatment of the trans isomer with ultraviolet irradiation. Anal. Sci. 2018, 34, 1195–1199. Available online: https://doi.org/10.2116/analsci.18P102 (accessed on 5 June 2019). [CrossRef] [PubMed]
- Matsunami, K.; Takamori, I.; Shinzato, T.; Aramoto, M.; Kondo, K.; Otsuka, K.; Takeda, Y. Radical-scavenging activities of new megastimane glucosides from Macaranga tanarius (L.) MULL.-ARG. Chem. Pharm. Bull. 2006, 54, 1403–1407. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: Samples of the compounds 1–4 are available from the authors. |


| Position | 1 * | 2 | 3 | 4 | Position | 1 * | 2 | 3 | 4 |
|---|---|---|---|---|---|---|---|---|---|
| 6 | 6.17 d (2.0) | 6.37 br s | 6.46 d (2.1) | 6.45 d (2.1) | 1′′′′ | - | 5.22 d (1.3) | 5.14 d (1.1) | 5.14 br s |
| 8 | 6.37 d (2.0) | 6.65 br s | 6.61 d (2.1) | 6.62 d (2.1) | 2′′′′ | 7.20 d (1.8) | 3.99 m | 3.90 m | 3.90 m |
| 2′ | 8.01 d (8.8) | 8.06 d (8.9) | 7.93 d (8.9) | 7.95 d (8.8) | 3′′′′ | - | 3.76 m | 3.69 m | 3.69 m |
| 3′ | 6.91 d (8.8) | 6.89 d (8.9) | 6.76 d (8.9) | 6.77 d (8.8) | 4′′′′ | - | 3.33 t (9.6) | 3.25 m | 3.42 m |
| 5′ | 6.91 d (8.8) | 6.89 d (8.9) | 6.76 d (8.9) | 6.77 d (8.8) | 5′′′′ | 6.83 d (8.2) | 4.02 dq (9.6, 6.18) | 3.93 dq (9.5, 6.2) | 3.68 dq (9.6, 6.2) |
| 6′ | 8.01 d (8.8) | 8.06 d (8.9) | 7.93 d (8.9) | 7.95 d (8.8) | 6′′′′ | 7.09 dd (8.2, 1.8) | 0.96 d (6.18) | 0.88 d (6.2) | 0.96 d (6.2) |
| 7′′′′ | 7.68 br d (15.9) | ||||||||
| 1′′ | 5.73 d (8.0) | 5.72 d (7.6) | 5.51 d (7.6) | 5.51 d (7.6) | 8′′′′ | 6.40 br d (15.9) | |||
| 2′′ | 5.23 dd (9.5, 8.0) | 3.60 m | 3.52 dd (9.6, 7.6) | 3.52 dd (9.6, 7.6) | OCH3 | 3.92 s | |||
| 3′′ | 3.91 t (9.5) | 3.55 t (9.0) | 3.45 m | 3.44 m | |||||
| 4′′ | 3.55 t (9.5) | 3.26 t (9.0) | 3.13 t (9.12) | 3.19 m | 1′′′′′ | 4.40 d (7.8) | 4.38 d (1.3) | 4.36 br s | |
| 5′′ | 3.39 ddd (9.5, 5.5, 2.0) | 3.31 m | 3.24 m | 3.24 m | 2′′′′′ | 3.20 dd (8.5, 7.8) | 3.38 m | 3.37 m | |
| 6′′ | 3.62 dd (12.0, 5.5) | 3.47 dd (12.0, 5.9) | 3.27 m | 3.27 m | 3′′′′′ | 3.31 m | 3.34 dd (9.4, 3.4) | 3.33 dd (9.4, 3.4) | |
| 3.81 dd (12.0, 2.0) | 3.71 dd (12.0, 2.1) | 3.71 m | 3.71 m | 4′′′′′ | 3.29 m | 3.12 t (9.4) | 3.25 m | ||
| 5′′′′′ | 3.22 m | 3.30 m | 3.25 m | ||||||
| 1′′′ | 4.43 d (7.8) | 5.29 d (7.8) | 4.98 d (7.4) | 4.98 d (7.3) | 6′′′′′ | 3.87 dd (11.5, 5.4) | 0.96 d (6.2) | 0.88 d (6.2) | |
| 2′′′ | 3.21 dd (8.8, 7.8) | 5.25 dd (9.0, 7.8) | 3.42 m | 3.41 t (9.4) | 3.64 dd (11.5, 1.3) | ||||
| 3′′′ | 3.31 t (8.8) | 3.92 t (9.0) | 3.43 m | 3.42 m | |||||
| 4′′′ | 3.28 t (8.8) | 3.76 m | 3.34 m | 3.34 t (9.4) | 2′′′′′′ | 7.55 d (8.6) | 6.98 d (1.7) | 7.25 d (8.6) | |
| 5′′′ | 3.34 m | 3.78 m | 3.72 m | 3.72 m | 3′′′′′′ | 6.67 d (8.6) | - | 6.66 d (8.6) | |
| 6′′′ | 3.62 dd (11.9, 2.2) | 3.75 dd (12.0, 5.1) | 4.17 dd (12.0, 2.0) | 4.18 dd (12.0, 2.1) | 5′′′′′′ | 6.67 d (8.6) | 6.67 d (8.2) | 6.66 d (8.6) | |
| 3.88 dd (11.9, 6.0) | 3.94 dd (12.0, 1.6) | 4.57 dd (12.0, 6.9) | 4.54 dd (12.0, 7.0) | 6′′′′′′ | 7.55 d (8.6) | 6.85 d (8.2, 1.7) | 7.25 d (8.6) | ||
| 7′′′′′′ | 6.86 br d (12.9) | 7.50 br d (15.8) | 7.50 br d (15.9) | ||||||
| 8′′′′′′ | 5.83 br d (12.9) | 6.28 br d (15.8) | 6.24 br d (15.9) | ||||||
| OCH3 | 3.75 s |
| Position | 1 * | 2 | 3 | 4 | Position | 1 * | 2 | 3 | 4 |
|---|---|---|---|---|---|---|---|---|---|
| 2 | 158.4 | 159.2 | 159.4 | 159.4 | 1′′′′ | 127.8 | 102.6 | 102.5 | 102.5 |
| 3 | 134.8 | 134.7 | 134.6 | 134.6 | 2′′′′ | 111.7 | 72.4 | 72.3 | 72.3 |
| 4 | 179.2 | 179.5 | 179.5 | 179.5 | 3′′′′ | 149.3 | 72.3 | 72.3 | 72.3 |
| 5 | 163.2 | 163.0 | 162.9 | 162.9 | 4′′′′ | 150.6 | 74.0 | 74.0 | 74.7 |
| 6 | 99.8 | 100.4 | 100.7 | 100.7 | 5′′′′ | 116.4 | 69.9 | 69.9 | 69.9 |
| 7 | 165.8 | 163.8 | 164.4 | 164.4 | 6′′′′ | 124.3 | 17.6 | 17.6 | 17.6 |
| 8 | 94.6 | 95.7 | 96.3 | 96.2 | 7′′′′ | 147.5 | |||
| 9 | 158.5 | 157.9 | 157.8 | 157.8 | 8′′′′ | 115.5 | |||
| 10 | 105.8 | 108.0 | 107.7 | 107.7 | 9′′′′ | 168.5 | |||
| 1′ | 122.7 | 122.8 | 122.8 | 122.8 | OCH3 | 56.4 | |||
| 2′ | 132.2 | 132.3 | 132.2 | 132.2 | |||||
| 3′ | 116.3 | 116.2 | 116.2 | 116.9 | 1′′′′′ | 105.0 | 102.2 | 102.2 | |
| 4′ | 161.6 | 161.6 | 161.4 | 161.5 | 2′′′′′ | 74.7 | 72.0 | 72.0 | |
| 5′ | 116.3 | 116.2 | 116.2 | 116.9 | 3′′′′′ | 77.8 | 72.0 | 72.0 | |
| 6′ | 132.2 | 132.3 | 132.2 | 132.2 | 4′′′′′ | 71.4 | 73.8 | 74.0 | |
| 5′′′′′ | 78.9 | 69.7 | 69.7 | ||||||
| 1′′ | 100.5 | 100.2 | 100.3 | 100.3 | 6′′′′′ | 62.5 | 17.8 | 17.8 | |
| 2′′ | 74.6 | 80.0 | 79.7 | 79.8 | |||||
| 3′′ | 84.9 | 78.9 | 79.0 | 79.0 | 1′′′′′′ | 127.6 | 127.6 | 127.1 | |
| 4′′ | 70.0 | 71.8 | 72.0 | 71.7 | 2′′′′′′ | 133.4 | 111.6 | 131.2 | |
| 5′′ | 78.4 | 78.1 | 77.2 | 77.2 | 3′′′′′′ | 115.9 | 149.2 | 116.2 | |
| 6′′ | 62.4 | 62.6 | 68.3 | 68.3 | 4′′′′′′ | 160.0 | 150.5 | 161.2 | |
| 5′′′′′′ | 115.9 | 116.5 | 116.2 | ||||||
| 1′′′ | 104.9 | 99.6 | 101.6 | 101.6 | 6′′′′′′ | 133.4 | 124.2 | 131.2 | |
| 2′′′ | 74.8 | 73.4 | 74.7 | 73.8 | 7′′′′′′ | 145.0 | 147.3 | 147.1 | |
| 3′′′ | 77.7 | 84.3 | 77.8 | 77.8 | 8′′′′′′ | 116.6 | 115.1 | 114.7 | |
| 4′′′ | 71.4 | 69.8 | 71.7 | 71.8 | 9′′′′′′ | 167.2 | 169.1 | 169.1 | |
| 5′′′ | 78.1 | 78.2 | 75.8 | 75.7 | OCH3 | 56.5 | |||
| 6′′′ | 62.5 | 62.2 | 64.6 | 64.6 |
| Isolated Compounds | DPPH (IC50, µM) | AGEs (IC50, µM) |
|---|---|---|
| Kaempferol 3-O-[2,6-di-O-α-l-rhamnopyranosyl]-β-d-glucoside (7) | >100 | 85.5 ± 3.5 |
| Myricetin 3-O-[2,6-di-O-α-l-rhamnopyranosyl]-β-d-glucoside (15) | 43.0 ± 1.15 | >100 |
| Myricetin 3-O-β-neohesperidoside (16) | 31.8 ± 0.63 | >100 |
| Myricetin 3-O-β-d-glucoside (17) | 14.5 ± 1.15 | 96.5 ± 1.8 |
| Myricetin (18) | 11.7 ± 1.8 | 34.9 ± 1.2 |
| Trolox | 29.2 ± 0.39 | n.d. |
| Aminoguanidine hydrochloride | n.d. | 7818 ± 34.4 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdulaziz Al-Hamoud, G.; Saud Orfali, R.; Sugimoto, S.; Yamano, Y.; Alothyqi, N.; Mohammed Alzahrani, A.; Matsunami, K. Four New Flavonoids Isolated from the Aerial Parts of Cadaba rotundifolia Forssk. (Qadab). Molecules 2019, 24, 2167. https://doi.org/10.3390/molecules24112167
Abdulaziz Al-Hamoud G, Saud Orfali R, Sugimoto S, Yamano Y, Alothyqi N, Mohammed Alzahrani A, Matsunami K. Four New Flavonoids Isolated from the Aerial Parts of Cadaba rotundifolia Forssk. (Qadab). Molecules. 2019; 24(11):2167. https://doi.org/10.3390/molecules24112167
Chicago/Turabian StyleAbdulaziz Al-Hamoud, Gadah, Raha Saud Orfali, Sachiko Sugimoto, Yoshi Yamano, Nafee Alothyqi, Ali Mohammed Alzahrani, and Katsuyoshi Matsunami. 2019. "Four New Flavonoids Isolated from the Aerial Parts of Cadaba rotundifolia Forssk. (Qadab)" Molecules 24, no. 11: 2167. https://doi.org/10.3390/molecules24112167
APA StyleAbdulaziz Al-Hamoud, G., Saud Orfali, R., Sugimoto, S., Yamano, Y., Alothyqi, N., Mohammed Alzahrani, A., & Matsunami, K. (2019). Four New Flavonoids Isolated from the Aerial Parts of Cadaba rotundifolia Forssk. (Qadab). Molecules, 24(11), 2167. https://doi.org/10.3390/molecules24112167