Identification of Quorum-Sensing Molecules of N-Acyl-Homoserine Lactone in Gluconacetobacter Strains by Liquid Chromatography-Tandem Mass Spectrometry
Abstract
:1. Introduction
2. Results
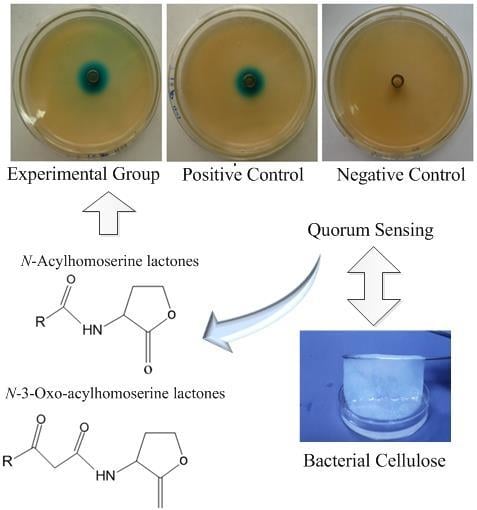
2.1. Detection of AHLs with a Biosensor System
2.2. Identification of AHLs with LC-MS/MS
2.3. Impact of GqqA Protein on the Biosynthesis of Cellulose in the Strain G. Xylinus CGMCC no. 2955
3. Discussion
4. Materials and Methods
4.1. Bacterial Strains and Culture Conditions
4.2. Biosensor System Method
4.3. LC-MS Method
4.4. Effects of Quorum-Quenching Protein GqqA on the Cellulose Production of CGMCC no. 2955
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Fuqua, W.C.; Winans, S.C.; Greenberg, E.P. Quorum sensing in bacteria: The LuxR-LuxI family of cell density-responsive transcriptional regulators. J. Bacteriol. 1994, 176, 269–275. [Google Scholar] [PubMed]
- Williams, P. Quorum sensing, communication and cross-kingdom signalling in the bacterial world. Microbiology 2007, 153, 3923–3938. [Google Scholar] [PubMed] [Green Version]
- De Kievit, T.R.; Iglewski, B.H. Bacterial quorum sensing in pathogenic relationships. Infect. Immun. 2000, 68, 4839–4849. [Google Scholar] [PubMed]
- Sturme, M.H.; Kleerebezem, M.; Nakayama, J.; Akkermans, A.D.; Vaughan, E.E.; de Vos, W.M. Cell to cell communication by autoinducing peptides in gram-positive bacteria. Antonie Van Leeuwenhoek 2002, 81, 233–243. [Google Scholar] [PubMed]
- Mok, K.C.; Wingreen, N.S.; Bassler, B.L. Vibrio harveyi quorum sensing: A coincidence detector for two autoinducers controls gene expression. Embo J. 2003, 22, 870–881. [Google Scholar] [PubMed]
- Engebrecht, J.; Nealson, K.; Silverman, M. Bacterial bioluminescence: Isolation and genetic analysis of functions from Vibrio fischeri. Cell 1983, 32, 773–781. [Google Scholar] [PubMed]
- Zhang, L.; Murphy, P.J.; Kerr, A.; Tate, M.E. Agrobacterium conjugation and gene regulation by N-acyl-l-homoserine lactones. Nature 1993, 362, 446–448. [Google Scholar]
- Zhu, J.; Mekalanos, J.J. Quorum sensing-dependent biofilms enhance colonization in Vibrio cholerae. Dev. Cell 2003, 5, 647–656. [Google Scholar]
- Whistler, C.A.; Pierson, L.S., III. Repression of phenazine antibiotic production in Pseudomonas aureofaciens strain 30–84 by RpeA. J. Bacteriol. 2003, 185, 3718–3725. [Google Scholar]
- von Bodman, S.B.; Bauer, W.D.; Coplin, D.L. Quorum sensing in plant-pathogenic bacteria. Annu. Rev. Phytopathol. 2003, 41, 455–482. [Google Scholar]
- Subramoni, S.; Florez Salcedo, D.V.; Suarez-Moreno, Z.R. A bioinformatic survey of distribution, conservation, and probable functions of LuxR solo regulators in bacteria. Front. Cell. Infect. Microbiol. 2015, 5, 16. [Google Scholar] [PubMed]
- Cleenwerck, I.; De Vos, P. Polyphasic taxonomy of acetic acid bacteria: An overview of the currently applied methodology. Int. J. Food Microbiol. 2008, 125, 2–14. [Google Scholar] [PubMed]
- Tanaka, M.; Murakami, S.; Shinke, R.; Aoki, K. Genetic characteristics of cellulose-forming acetic acid bacteria identified phenotypically as Gluconacetobacter xylinus. Biosci. Biotechnol. Biochem. 2000, 64, 757–760. [Google Scholar] [PubMed]
- McLean, R.J.; Pierson, L.S.; Fuqua, C. A simple screening protocol for the identification of quorum signal antagonists. J. Microbiol. Methods 2004, 58, 351–360. [Google Scholar] [PubMed]
- Erdönmez, D.; Rad, A.Y.; Aksöz, N. Quorum sensing molecules production by nosocomial and soil isolates Acinetobacter baumannii. Arch. Microbiol. 2017, 199, 1325–1334. [Google Scholar]
- Wang, J.; Ding, L.; Li, K.; Schmieder, W.; Geng, J.; Xu, K.; Zhang, Y.; Ren, H. Development of an extraction method and LC–MS analysis for N-acylated-l-homoserine lactones (AHLs) in wastewater treatment biofilms. J. Chromatogr. B 2017, 1041, 37–44. [Google Scholar]
- Morin, D.; Grasland, B.; Valléeréhel, K.; Dufau, C.; Haras, D. On-line high-performance liquid chromatography-mass spectrometric detection and quantification of N-acylhomoserine lactones, quorum sensing signal molecules, in the presence of biological matrices. J. Chromatogr. A 2003, 1002, 79. [Google Scholar]
- Bruhn, J.B.; Christensen, A.B.; Flodgaard, L.R.; Nielsen, K.F.; Larsen, T.O.; Givskov, M.; Gram, L. Presence of acylated homoserine lactones (AHLs) and AHL-producing bacteria in meat and potential role of AHL in spoilage of meat. Appl. Environ. Microbiol. 2004, 70, 4293–4302. [Google Scholar]
- Fletcher, M.; Cámara, M.; Barrett, D.A.; Williams, P. Biosensors for qualitative and semiquantitative analysis of quorum sensing signal molecules. Pseudomonas Methods Protoc. 2014, 1149, 245–254. [Google Scholar]
- Jose Valera, M.; Mas, A.; Streit, W.R.; Mateo, E. GqqA, a novel protein in Komagataeibacter europaeus involved in bacterial quorum quenching and cellulose formation. Microb. Cell. Fact. 2016, 15, 88. [Google Scholar]
- Iida, A.; Ohnishi, Y.; Horinouchi, S. Control of acetic acid fermentation by quorum sensing via N-acylhomoserine lactones in Gluconacetobacter intermedius. J. Bacteriol. 2008, 190, 2546–2555. [Google Scholar] [PubMed]
- Iida, A.; Ohnishi, Y.; Horinouchi, S. An OmpA family protein, a target of the GinI/GinR quorum-sensing system in Gluconacetobacter intermedius, controls acetic acid fermentation. J. Bacteriol. 2008, 190, 5009–5019. [Google Scholar] [PubMed]
- Iida, A.; Ohnishi, Y.; Horinouchi, S. Identification and characterization of target genes of the GinI/GinR quorum-sensing system in Gluconacetobacter intermedius. Microbiology 2009, 155, 3021–3032. [Google Scholar]
- Weinhouse, H.; Sapir, S.; Amikam, D.; Shilo, Y.; Volman, G.; Ohana, P.; Benziman, M. c-di-GMP-binding protein, a new factor regulating cellulose synthesis in Acetobacter xylinum. Febs Lett. 1997, 416, 207–211. [Google Scholar] [PubMed]
- Tonouchi, N.; Tsuchida, T.; Yoshinaga, F.; Beppu, T.; Horinouchi, S. Characterization of the biosynthetic pathway of cellulose from glucose and fructose in Acetobacter xylinum. Biosci. Biotechnol. Biochem. 1996, 60, 1377–1379. [Google Scholar]
- Ross, P.; Weinhouse, H.; Aloni, Y.; Michaeli, D.; Weinberger-Ohana, P.; Mayer, R.; Braun, S.; De Vroom, E.; Van der Marel, G.; Van Boom, J. Regulation of cellulose synthesis in Acetobacter xylinum by cyclic diguanylic acid. Nature 1987, 325, 279–281. [Google Scholar] [PubMed]
- Du, X.-J.; Jia, S.-R.; Yang, Y.; Wang, S. Genome sequence of Gluconacetobacter sp. strain SXCC-1, isolated from Chinese vinegar fermentation starter. J. Bacteriol. 2011, 193, 3395–3396. [Google Scholar]
- Fuqua, C.; Winans, S.C. Conserved cis-acting promoter elements are required for density-dependent transcription of Agrobacterium tumefaciens conjugal transfer genes. J. Bacteriol. 1996, 178, 435–440. [Google Scholar]
- Pechere, J.C. Azithromycin reduces the production of virulence factors in Pseudomonas aeruginosa by inhibiting quorum sensing. Jpn. J. Antibiot. 2001, 54 Suppl. C, 87–89. [Google Scholar]
- Lee, J.; Zhang, L. The hierarchy quorum sensing network in Pseudomonas Aeruginosa. Protein Cell 2015, 6, 26–41. [Google Scholar]
- Yeon, K.-M.; Cheong, W.-S.; Oh, H.-S.; Lee, W.-N.; Hwang, B.-K.; Lee, C.-H.; Beyenal, H.; Lewandowski, Z. Quorum sensing: A new biofouling control paradigm in a membrane bioreactor for advanced wastewater treatment. Environ. Sci. Technol. 2009, 43, 380–385. [Google Scholar] [PubMed]
- Ravn, L.; Christensen, A.B.; Molin, S.; Givskov, M.; Gram, L. Methods for detecting acylated homoserine lactones produced by Gram-negative bacteria and their application in studies of AHL-production kinetics. J. Microbiol. Methods 2001, 44, 239–251. [Google Scholar] [PubMed]
- Behrend, O.; Ax, K.; Schubert, H. Influence of continuous phase viscosity on emulsification by ultrasound. Ultrason. Sonochem. 2000, 7, 77–85. [Google Scholar] [PubMed]
- Ortori, C.A.; Halliday, N.; Cámara, M.; Williams, P.; Barrett, D.A. LC-MS/MS quantitative analysis of quorum sensing signal molecules. Pseudomonas Methods Protoc. 2014, 1149, 255–270. [Google Scholar]
Sample Availability: Sample of the compound bacterial cellulose is available from the authors. |




| No. | Abbreviation | Precursor Ion (m/z) | Peak Intensity | Retention Time (min) |
|---|---|---|---|---|
| 1 | C2-HSL | 144 | 1.04 × 104 | 1.91 |
| 2 | C4-HSL | 172 | 3.90 × 104 | 2.54 |
| 3 | C6-HSL | 200 | 4.78 × 103 | 8.30 |
| 4 | 3-oxo-C8-HSL | 242 | 2.07 × 103 | 12.95 |
| 5 | C10-HSL | 256 | 2.08 × 106 | 19.46 |
| 6 | C12-HSL | 284 | 3.03 × 104 | 23.81 |
| 7 | C14-HSL | 312 | 1.60 × 104 | 29.54 |
| 8 | C2-HSL | 144 | 4.74 × 103 | 2.11 |
| 9 | C4-HSL | 172 | 5.80 × 104 | 2.68 |
| 10 | C6-HSL | 200 | 1.53 × 103 | 8.19 |
| 11 | 3-oxo-C10-HSL | 270 | 8.04 × 105 | 17.23 |
| 12 | C12-HSL | 284 | 4.79 × 104 | 23.77 |
| 13 | C14-HSL | 312 | 6.70 × 103 | 29.46 |
| 14 | C2-HSL | 144 | 5.49 × 106 | 1.90 |
| 15 | C4-HSL | 172 | 8.07 × 107 | 2.56 |
| 16 | C6-HSL | 200 | 1.32 × 106 | 8.00 |
| 17 | C10-HSL | 256 | 4.18 × 106 | 22.33 |
| 18 | C12-HSL | 284 | 1.68 × 106 | 24.59 |
| 19 | 3-oxo-C12-HSL | 298 | 8.92 × 104 | 26.18 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Liu, L.-P.; Huang, L.-H.; Ding, X.-T.; Yan, L.; Jia, S.-R.; Dai, Y.-J.; Xie, Y.-Y.; Zhong, C. Identification of Quorum-Sensing Molecules of N-Acyl-Homoserine Lactone in Gluconacetobacter Strains by Liquid Chromatography-Tandem Mass Spectrometry. Molecules 2019, 24, 2694. https://doi.org/10.3390/molecules24152694
Liu L-P, Huang L-H, Ding X-T, Yan L, Jia S-R, Dai Y-J, Xie Y-Y, Zhong C. Identification of Quorum-Sensing Molecules of N-Acyl-Homoserine Lactone in Gluconacetobacter Strains by Liquid Chromatography-Tandem Mass Spectrometry. Molecules. 2019; 24(15):2694. https://doi.org/10.3390/molecules24152694
Chicago/Turabian StyleLiu, Ling-Pu, Long-Hui Huang, Xiao-Tong Ding, Lin Yan, Shi-Ru Jia, Yu-Jie Dai, Yan-Yan Xie, and Cheng Zhong. 2019. "Identification of Quorum-Sensing Molecules of N-Acyl-Homoserine Lactone in Gluconacetobacter Strains by Liquid Chromatography-Tandem Mass Spectrometry" Molecules 24, no. 15: 2694. https://doi.org/10.3390/molecules24152694
APA StyleLiu, L. -P., Huang, L. -H., Ding, X. -T., Yan, L., Jia, S. -R., Dai, Y. -J., Xie, Y. -Y., & Zhong, C. (2019). Identification of Quorum-Sensing Molecules of N-Acyl-Homoserine Lactone in Gluconacetobacter Strains by Liquid Chromatography-Tandem Mass Spectrometry. Molecules, 24(15), 2694. https://doi.org/10.3390/molecules24152694