Fungicide Tebuconazole Influences the Structure of Human Serum Albumin Molecule
Abstract
:1. Introduction
2. Results and Discussion
2.1. Differential Scanning Calorimetry
2.2. Fluorescence Quenching
2.3. Determination of Binding Parameters
2.4. Thermodynamic Parameters
2.5. Synchronous Fluorescence Spectroscopy
2.6. Site Markers Competitive Experiments
2.7. 3D Fluorescence Spectroscopy
2.8. Circular Dichroism
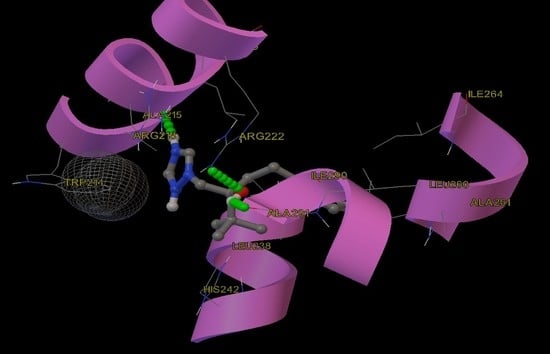
2.9. Molecular Docking
3. Materials and Methods
3.1. Chemicals and Reagents
3.2. Differential Scanning Calorimetry
3.3. Fluorescence Measurements
3.4. Circular Dichroism (CD) Studies
3.5. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Clausen, C.A.; Yang, V. Protecting wood from mould, decay, and termites with multi-component biocide systems. Int. Biodeterior. Biodegrad. 2007, 59, 20–24. [Google Scholar] [CrossRef] [Green Version]
- Fera, M.T.; La camera, E.; Desarro, A. New triazoles and echinocandins: Mode of action, in itro activity and mechanisms of resistance. Expert Rev. Anti-infect. Ther. 2009, 7, 981–998. [Google Scholar] [CrossRef] [PubMed]
- U.S. Environmental Protection Agency. Available online: https://www.epa.gov/pesticide-science-and-assessing-pesticide-risks/evaluating-pesticides-carcinogenic-potential#a (accessed on 10 January 2014).
- EFSA. Available online: https://www.efsa.europa.eu/en/efsajournal/pub/3485 (accessed on 3 January 2014).
- Muñoz-Leoz, B.; Ruiz-Romera, E.; Antigüedad, I.; Garbisu, C. Tebuconazole application decreases soil microbial biomass and activity. Soil Biol. Biochem. 2011, 43, 2176–2183. [Google Scholar] [CrossRef]
- Bayer crop science limited. Environmental information sheet Folicur® MAPP number 11278. Available online: https://voluntaryinitiative.org.uk/media/1379/folicur-version-3-guideline-version-4.pdf (accessed on 15 February 2015).
- Montuelle, B.; Dorigo, U.; Bérard, A.; Volat, B.; Bouchez, A.; Tlili, A.; Gouy, S.; Pesce, S. The periphyton as a multimetricbioindicator for assessing the impact of land use on rivers: An overview of the Ardières-Morcille experimental watershed (France). In Global Change and River Ecosystems—Implications for Structure, Function and Ecosystem Services; Stevenson, R.J., Sabater, S., Eds.; Springer: Dordrecht, The Netherlands, 2010; pp. 123–141. [Google Scholar]
- Elsaesser, D.; Schulz, R. Mitigation of fungicide pollution in vegetated agricultural surface waters: GIS modelling and monitoring in the field. In Proceedings of the SETAC Europe 18th Annual Meeting, Warsaw, Poland, 25–29 May 2008; pp. 406–407. [Google Scholar]
- Yu, L.; Chen, M.; Liu, Y.; Gui, W.; Zhu, G. Thyroid endocrine disruption in zebrafish larvae following exposure to hexaconazole and tebuconazole. Aquat. Toxicol. 2013, 138, 35–42. [Google Scholar] [CrossRef] [PubMed]
- Holečková, B.; Šiviková, K.; Dianovský, J.; Galdíková, M. Effect of triazole pesticide formulation on bovine culture cells. J. Environ. Sci. Health B 2013, 48, 1080–1088. [Google Scholar] [CrossRef] [PubMed]
- Fustinoni, S.; Polledri, E.; Mercadante, R.; Rubino, F.M.; Colosio, C.; Moretto, A. Time course of excretion of tebuconazole and its metabolites in vineyard workers. G. Ital. Med. Lav. 2012, 34, 423–424. [Google Scholar]
- Schummer, C.; Salquebre, G.; Briand, O.; Millet, M.; Appenzeller, B.M. Determination of farm workers′ exposure to pesticides by hair analysis. Toxicol. Lett. 2012, 210, 203–210. [Google Scholar] [CrossRef]
- Mas, N.; Galiana, I.; Hurtado, S.; Mondragón, L.; Bernardos, A.; Sancenón, F.; Murguía, J.R. Enhanced antifungal efficacy of tebuconazole using gated ph-driven mesoporous nanoparticles. Int. J. Nanomed. 2014, 9, 2597. [Google Scholar]
- Monkos, K. On the hydrodynamics and temperature dependence of the solution conformation of human serum albumin from viscometry approach. Biochim. Biophys. Acta 2004, 1700, 27–34. [Google Scholar] [CrossRef]
- Peters, T.J. The albumin molecule: Its structure and chemical properties. In All about Albumin: Biochemistry, Genetics and Medical Applications; Peters, T.J., Ed.; Academic Press: San Diego, CA, USA, 1996; pp. 9–75. [Google Scholar]
- Fehske, K.J.; Muller, W.E.; Wollert, U. The location of drug binding sites in human serum albumin. Biochem. Pharmacol. 1981, 30, 687–692. [Google Scholar] [CrossRef]
- Kragh-Hansen, U. Molecular aspects of ligand binding to serum albumin. Pharmacol. Rev. 1981, 33, 17–53. [Google Scholar] [PubMed]
- Putnam, F.W.; Lozier, J.; Takahashi, N. Complete amino acid sequence of human plasma beta 2-glycoprotein I. Proc. Natl. Acad. Sci. USA 1984, 81, 3640–3644. [Google Scholar]
- Miškovský, P. Hypericin—A new antiviral and antitumor photosensitizer: Mechanism of action and interaction with biological macromolecules. Curr. Drug Targets 2002, 3, 55–84. [Google Scholar] [CrossRef] [PubMed]
- Fabriciová, G.; Sanchez-Cortes, S.; Garcia-Ramos, J.V.; Miškovský, P. Surface-enhanced Raman spectroscopy study of the interaction of the antitumoral drug emodin with human serum albumin. Biopolymers 2004, 74, 125–130. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Silva, D.; Cortez, C.M.; Cunha-Bastos, J.; Louro, S.R. Methyl parathion interaction with human and bovine serum albumin. Toxicol. Lett. 2004, 147, 53–61. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Guo, J.; Xu, B.; Chen, Z. Binding of chlorpyrifos and cypermethrin to blood proteins. Pestic. Biochem. Physiol. 2006, 85, 110–114. [Google Scholar] [CrossRef]
- Yan, C.N.; Mei, P.; Gua, Z.J.; Liu, Y. Studies on thermodynamics features of the interaction between imidacloprid and bovine serum albumin. Chin. J. Chem. 2007, 25, 1085–1089. [Google Scholar] [CrossRef]
- Wang, Y.Q.; Tang, B.P.; Zhang, H.M.; Zhou, Q.H.; Zhang, G.C. Studies on the interaction between imidacloprid and human serum albumin: Spectroscopic approach. J. Photochem. Photobiol. B 2009, 94, 183–190. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, X.; Wang, J.; Zhao, Y.; He, W.; Guo, Z. Noncovalent interactions between a trinuclearmonofunctional platinum complex and human serum albumin. Inorg. Chem. 2011, 50, 12661–12668. [Google Scholar] [CrossRef]
- Zhang, J.; Zhuang, S.; Tong, C.; Liu, W. Probing the molecular interaction of triazole fungicides with human serum albumin by multispectroscopic techniques and molecular modeling. J. Agric. Food Chem. 2013, 61, 7203–7211. [Google Scholar] [CrossRef]
- Sułkowska, A.; Maciążek-Jurczyk, M.; Bojko, B.; Rownicka, J.; Zubik-Skupień, I.; Temba, E.; Sułkowski, W.W. Competitive binding of phenylbutazone and colchicine to serum albumin in multidrug therapy: A spectroscopic study. J. Mol. Struct. 2008, 881, 97–106. [Google Scholar] [CrossRef]
- Staničová, J.; Želonková, K.; Verebová, V.; Holečková, B.; Dianovský, J. Interaction of the Fungicide Tebuconazole with Human Serum Albumin: A Preliminary Study. Folia Veterinaria 2018, 62, 85–91. [Google Scholar] [CrossRef] [Green Version]
- Shrake, A.; Ross, P.D. Ligand-induced biphasic protein denaturation. J. Biol. Chem. 1990, 265, 5055–5059. [Google Scholar] [PubMed]
- Faroongsarng, D. Assessment of the dissociation energetics of some selected ligand drugs bound on human serum albumin by differential scanning calorimetry. AAPS PharmSciTech 2016, 17, 474–481. [Google Scholar] [CrossRef] [PubMed]
- Celej, M.S.; Dassie, S.A.; González, M.; Bianconi, M.L.; Fidelio, G.D. Differential scanning calorimetry as a tool to estimate binding parameters in multiligand binding proteins. Anal. Biochem. 2006, 350, 277–284. [Google Scholar] [CrossRef] [PubMed]
- Koslen, M.M.; Eskew, M.W.; Pinkert, V.; Hoang, H.; Manyanga, F.; Dean, W.L.; Benight, A.S. Capture Reagent and Strategy for Retrieving Albumin-Bound Ligands from Plasma. Adv. Biol. Chem. 2019, 9, 110–134. [Google Scholar] [CrossRef] [Green Version]
- Michnik, A.; Michalik, K.; Kluczewska, A.; Drzazga, Z. Comparative DSC study of human and bovine serum albumin. J. Therm. Anal. Calorim. 2005, 84, 113–117. [Google Scholar] [CrossRef]
- Shrake, A.; Ross, P.D. Origins and consequences of ligand-induced multiphasic thermal protein denaturation. Biopolymers 1992, 32, 925–940. [Google Scholar] [CrossRef]
- Lemli, B.; Derdák, D.; Laczay, P.; Kovács, D.; Kunsági-Máté, S. Noncovalent Interaction of Tilmicosin with Bovine Serum Albumin. Molecules 2018, 23, 1915. [Google Scholar] [CrossRef]
- Lakowicz, J.R. Quenching of fluorescence. In Principles of Fluorescence Spectroscopy., 3rd ed.; Lakowicz, J.R., Ed.; Plenum Press: New York, NY, USA, 1983; pp. 199–227. [Google Scholar]
- Zhang, G.; Wang, L.; Pan, J. Probing the binding of the flavonoid diosmetin to human serum albumin by multispectroscopic techniques. J. Agric. Food Chem. 2012, 60, 2721–2729. [Google Scholar] [CrossRef]
- Lu, Y.; Feng, Q.; Cui, F.; Xing, W.; Zhang, G.; Yao, X. Interaction of 3-azido-3-deamino daunorubicin with human serum albumin: Investigation by fluorescence spectroscopy and molecular modelling methods. Bioorg. Med. Chem. Lett. 2010, 20, 6899–6904. [Google Scholar] [CrossRef] [PubMed]
- Eftink, M.R.; Ghiron, C.A. Fluorescence quenching studies with proteins. Anal. Biochem. 1981, 114, 199–227. [Google Scholar] [CrossRef]
- Lakowicz, J.R.; Weber, G. Quenching of fluorescence by oxygen. Probe for structural fluctuations in macromolecules. Biochemistry 1973, 12, 4161–4170. [Google Scholar] [CrossRef] [PubMed]
- Jiang, M.; Xie, M.-X.; Zheng, D.; Liu, Y.; Li, X.-Y.; Chen, X. Spectroscopic studies on the interaction of cinnamic acid and its hydroxyl derivatives with human serum albumin. J. Mol. Struct. 2004, 692, 71–80. [Google Scholar]
- Huang, Y.; Zhang, Z.; Zhang, D.; Lv, J. Flow-injection analysis chemiluminescence detection combined with microdialysis sampling for studying protein binding of drug. Talanta 2001, 53, 835–841. [Google Scholar] [CrossRef]
- Tian, J.; Liu, J.; He, W.; Hu, Z.; Yao, X.; Chen, X. Probing the binding of scutellarin to human serum albumin by circular dichroism, fluorescence spectroscopy, FTIR, and molecular modelling method. Biomacromolecules 2004, 5, 1956–1961. [Google Scholar] [CrossRef] [PubMed]
- Ross, P.D.; Subramanian, S. Thermodynamics of protein association reactions: Forces contributing to stability. Biochemistry 1981, 20, 3096–3102. [Google Scholar] [CrossRef] [PubMed]
- Némethy, G.; Scheraga, H.A. The structure of water and hydrophobic bonding in proteins. iii. The thermodynamic properties of hydrophobic bonds in proteins1, 2. J. Phys. Chem. A 1962, 66, 1773–1789. [Google Scholar] [CrossRef]
- Pan, X.; Qin, P.; Liu, R.; Wang, J. Characterizing the interaction between tartrazine and two serum albumins by a hybrid spectroscopic approach. J. Agric. Food Chem. 2011, 59, 6650–6656. [Google Scholar] [CrossRef] [PubMed]
- Xiang, G.; Tong, C.; Lin, H. Nitroaniline isomers interaction with bovine serum albumin and toxicological implications. J. Fluoresc. 2007, 17, 512–521. [Google Scholar] [CrossRef] [PubMed]
- Vekshin, N.L. Division of tyrosine and tryptophan fluorescent components by synchronous scanning method. Biofizika 1996, 41, 1176–1179. [Google Scholar] [PubMed]
- Congdon, R.W.; Muth, G.W.; Splittgerber, A.G. The binding interaction of Coomassie blue with proteins. Anal. Biochem. 1993, 213, 407–413. [Google Scholar] [CrossRef] [PubMed]
- Deeb, O.; Rosales-Hernández, M.C.; Gómez-Castro, C.; Garduno-Juárez, R.; Correa-Basurto, J. Exploration of human serum albumin binding sites by docking and molecular dynamics flexible ligand–protein interactions. Biopolymers 2010, 93, 161–170. [Google Scholar] [CrossRef] [PubMed]
- He, X.M.; Carter, D.C. Atomic structure and chemistry of human serum albumin. Nature 1992, 358, 209. [Google Scholar] [CrossRef] [PubMed]
- Xu, T.; Guo, X.; Zhang, L.; Pan, F.; Lv, J.; Zhang, Y.; Jin, H. Multiple spectroscopic studies on the interaction between olaquindox, a feed additive, and bovine serum albumin. Food Chem. Toxicol. 2012, 50, 2540–2546. [Google Scholar] [CrossRef] [PubMed]
- Sudlow, G.; Birkett, D.J.; Wade, D.N. Spectroscopic techniques in the study of protein binding. A fluorescence technique for the evaluation of the albumin binding and displacement of warfarin and warfarin-alcohol. Clin. Exp. Pharmacol. Physiol. 1975, 2, 129–140. [Google Scholar] [CrossRef] [PubMed]
- Rodriguez-Cuesta, M.J.; Boqué, R.; Rius, F.X.; Zamora, D.P.; Galera, M.M.; Frenich, A.G. Determination of carbendazim, fuberidazole and thiabendazole by three-dimensional excitation–emission matrix fluorescence and parallel factor analysis. Anal. Chim. Acta 2003, 491, 47–56. [Google Scholar] [CrossRef]
- Trevisan, M.G.; Poppi, R.J. Determination of doxorubicin in human plasma by excitation–emission matrix fluorescence and multi-way analysis. Anal. Chim. Acta 2003, 493, 69–81. [Google Scholar] [CrossRef]
- Kalaiarasi, G.; Rajkumar, S.R.J.; Dharani, S.; Malecko, J.G.; Prabhakaran, R. An investigation on 3-acetyl-7-methoxy-coumarin Schiff bases and their Ru (II) metallates with potent antiproliferative activity and enhanced LDH and NO release. RSC Adv. 2018, 8, 1539–1561. [Google Scholar] [CrossRef]
- Tian, F.F.; Jiang, F.L.; Han, X.L.; Xiang, C.; Ge, Y.S.; Li, J.H.; Zhang, Y.; Li, R.; Ding, X.L.; Liu, Y. Synthesis of a novel hydrazone derivative and biophysical studies of its interactions with bovine serum albumin by spectroscopic, electrochemical, and molecular docking methods. J. Phys. Chem. B 2010, 114, 14842–14853. [Google Scholar] [CrossRef]
- He, Y.; Wang, Y.; Tang, L.; Liu, H.; Chen, W.; Zheng, Z.; Zou, G. Binding of puerarin to human serum albumin: A spectroscopic analysis and molecular docking. J. Fluoresc. 2008, 18, 433–442. [Google Scholar] [CrossRef] [PubMed]
- Kandagal, P.B.; Ashoka, S.; Seetharamappa, J.; Shaikh, S.M.T.; Jadegoud, Y.; Ijare, O.B. Study of the interaction of an anticancer drug with human and bovine serum albumin: Spectroscopic approach. J. Pharm. Biomed. Anal. 2006, 41, 393–399. [Google Scholar] [CrossRef] [PubMed]
- Yang, P.; Gao, F. The principle of bioinorganic chemistry; Science Press: Beijing, China, 2002; p. 349. [Google Scholar]
- Lu, Z.X.; Cui, T.; Shi, Q.L. Applications of circular dichroism and optical rotatory dispersion in molecular biology; Science Press: Beijing, China, 1987; pp. 79–82. [Google Scholar]
- Ding, F.; Diao, J.X.; Sun, Y.; Sun, Y. Bioevaluation of human serum albumin–hesperidin bioconjugate: Insight into protein vector function and conformation. J. Agric. Food Chem. 2012, 60, 7218–7228. [Google Scholar] [CrossRef] [PubMed]
- Greenfield, N.J. Using circular dichroism spectra to estimate protein secondary structure. Nat. Protoc. 2006, 1, 2876. [Google Scholar] [CrossRef] [PubMed]
- Trott, O.; Olson, A.J. AutoDockVina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading. J.comput.chem. 2010, 31, 455–461. [Google Scholar] [PubMed]
- Evoli, S.; Mobley, D.L.; Guzzi, R.; Rizzuti, B. Multiple binding modes of ibuprofen in human serum albumin identified by absolute binding free energy calculations. Phys. Chem. Chem. Phys. 2016, 18, 32358–32368. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ghuman, J.; Zunszain, P.A.; Petitpas, I.; Bhattacharya, A.A.; Otagiri, M.; Curry, S. Structural basis of the drug-binding specificity of human serum albumin. J. Mol. Biol. 2005, 353, 38–52. [Google Scholar] [CrossRef] [PubMed]
- Sugio, S.; Kashima, A.; Mochizuki, S.; Noda, M.; Kobayashi, K. Crystal structure of human serum albumin at 2.5 Å resolution. Protein Eng. 1999, 12, 439–446. [Google Scholar] [CrossRef]
- Lee, P.; Wu, X. Modifications of human serum albumin and their binding effect. Curr. Pharm. Des. 2015, 21, 1862–1865. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are available from the authors and commercially. |





| Ratio TB/HSA | tD1 (°C) * | tD2 (°C) + | ΔH (kJ/mol) |
|---|---|---|---|
| 0 | 57.20 | 65.53 | 527.97 |
| 1/1 | 57.86 | 67.08 | 604.75 |
| 5/1 | 59.67 | 71.57 | 652.22 |
| 7/1 | 60.46 | 72.93 | 710.02 |
| 10/1 | 60.49 | 72.99 | 730.66 |
| Temperature (°C) | KSV (L/mol) | KA (L/mol) | n | ΔG (kJ/mol) | ΔH (kJ/mol) | ΔS (J/mol·K) |
|---|---|---|---|---|---|---|
| 25 * | 7.26 × 103 ± 0.09 | 8.51 × 103 ± 0.06 | 1.01 ± 0.02 | −22.68 ± 0.001 | −56.96 ± 0.02 | −115.98 ± 0.07 |
| 30 | 4.77 × 103 ± 0.02 | 4.85 × 103 ± 0.02 | 0.98 ± 0.02 | −22.10 ± 0.001 | ||
| 37 | 3.15 × 103 ± 0.02 | 3.46 × 103 ± 0.09 | 0.96 ± 0.02 | −20.99 ± 0.002 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Želonková, K.; Havadej, S.; Verebová, V.; Holečková, B.; Uličný, J.; Staničová, J. Fungicide Tebuconazole Influences the Structure of Human Serum Albumin Molecule. Molecules 2019, 24, 3190. https://doi.org/10.3390/molecules24173190
Želonková K, Havadej S, Verebová V, Holečková B, Uličný J, Staničová J. Fungicide Tebuconazole Influences the Structure of Human Serum Albumin Molecule. Molecules. 2019; 24(17):3190. https://doi.org/10.3390/molecules24173190
Chicago/Turabian StyleŽelonková, Katarína, Samuel Havadej, Valéria Verebová, Beáta Holečková, Jozef Uličný, and Jana Staničová. 2019. "Fungicide Tebuconazole Influences the Structure of Human Serum Albumin Molecule" Molecules 24, no. 17: 3190. https://doi.org/10.3390/molecules24173190
APA StyleŽelonková, K., Havadej, S., Verebová, V., Holečková, B., Uličný, J., & Staničová, J. (2019). Fungicide Tebuconazole Influences the Structure of Human Serum Albumin Molecule. Molecules, 24(17), 3190. https://doi.org/10.3390/molecules24173190