Data-Dependent Acquisition and Database-Driven Efficient Peak Annotation for the Comprehensive Profiling and Characterization of the Multicomponents from Compound Xueshuantong Capsule by UHPLC/IM-QTOF-MS
Abstract
:1. Introduction
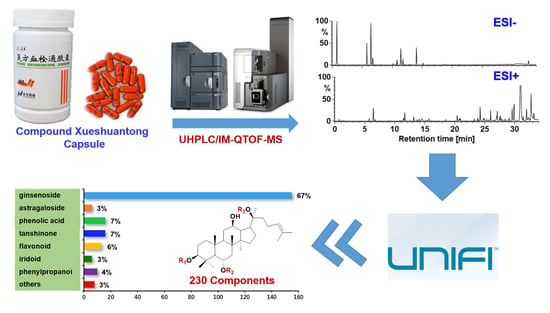
2. Results and Discussion
2.1. Optimization of a UHPLC/IM-QTOF-MS Approach Dedicated to Separating and Characterizing the Multicomponents from CXC
2.2. Development of a “Component Knockout” Approach to Enhance the Exposure of Minor Components from CXC
2.3. Systematic Characterization of Multiple Types of Components from CXC by Applying UNIFITM to Annotate the DDA Data Obtained in Both the Negative and Positive ESI Modes
2.3.1. Characterization of Ginsenosides
2.3.2. Characterization of Astragalosides
2.3.3. Characterization of Phenolic Acids
2.3.4. Characterization of Tanshinones
2.3.5. Characterization of Flavonoids
2.3.6. Characterization of Iridoids
2.3.7. Characterization of Phenylpropanoids
3. Materials and Methods
3.1. Reagents and Chemicals
3.2. Sample Preparation
3.3. UHPLC/IM-QTOF-MS
3.4. Date Processing
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Yang, W.Z.; Zhang, Y.B.; Wu, W.Y.; Huang, L.Q.; Guo, D.A.; Liu, C.X. Approaches to establish Q-markers for the quality standards of traditional Chinese medicine. Acta. Pharm. Sin. B 2017, 7, 439–446. [Google Scholar] [CrossRef] [PubMed]
- Yan, Y.; Song, Q.Q.; Chen, X.J.; Li, J.; Li, P.; Wang, Y.T.; Liu, T.X.; Song, Y.L.; Tu, P.F. Simultaneous determination of components with wide polarity and content ranges in Cistanche tubulosa using serially coupled reverse phase-hydrophilic interaction chromatography-tandem mass spectrometry. J. Chromatogr. A 2017, 1501, 39–50. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.L.; Yang, W.Z.; Wu, W.Y.; Da, J.; Hou, J.J.; Zhang, J.X.; Zhang, Y.H.; Jin, Y.; Yang, M.; Jiang, B.H.; et al. Simultaneous quantitation of five Panax notoginseng saponins by multi heart-cutting two-dimensional liquid chromatography: Method development and application to the quality control of eight Notoginseng containing Chinese patent medicines. J. Chromatogr. A 2015, 1402, 71–81. [Google Scholar] [CrossRef] [PubMed]
- Ganzera, M.; Sturm, S. Recent advances on HPLC/MS in medicinal plant analysis—An update covering 2011–2016. J. Pharm. Biomed. Anal. 2018, 147, 211–233. [Google Scholar] [CrossRef] [PubMed]
- Yao, C.L.; Yang, W.Z.; Si, W.; Shen, Y.; Zhang, N.X.; Chen, H.L.; Pan, H.Q.; Yang, M.; Wu, W.Y.; Guo, D.A. An enhanced targeted identification strategy for the selective identification of flavonoid O-glycosides from Carthamus tinctorius by integrating offline two-dimensional liquid chromatography/linear ion-trap-Orbitrap mass spectrometry, high-resolution diagnostic product ions/neutral loss filtering and liquid chromatography-solid phase extraction-nuclear magnetic resonance. J. Chromatogr. A 2017, 1491, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Fekete, S.; Veuthey, J.L.; Guillarme, D. Comparison of the most recent chromatographic approaches applied for fast and high resolution separations: Theory and practice. J. Chromatogr. A 2015, 1408, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Pirok, B.W.J.; Stoll, D.R.; Schoenmakers, P.J. Recent developments in two-dimensional liquid chromatography: Fundamental improvements for practical applications. Anal. Chem. 2019, 91, 240–263. [Google Scholar] [CrossRef] [PubMed]
- Ji, S.; Wang, S.; Xu, H.S.; Su, Z.Y.; Tang, D.Q.; Qiao, X.; Ye, M. The application of on-line two-dimensional liquid chromatography (2DLC) in the chemical analysis of herbal medicines. J. Pharm. Biomed. Anal. 2018, 160, 301–313. [Google Scholar] [CrossRef]
- Zhang, A.H.; Sun, H.; Yan, G.L.; Wang, X.J. Recent developments and emerging trends of mass spectrometry for herbal ingredients analysis. Trends Anal. Chem. 2017, 94, 70–76. [Google Scholar] [CrossRef]
- Fu, L.L.; Ding, H.; Han, L.F.; Jia, L.; Yang, W.Z.; Zhang, C.X.; Hu, Y.; Zuo, T.T.; Gao, X.M.; Guo, D.A. Simultaneously targeted and untargeted multicomponent characterization of Erzhi Pill by offline two-dimensional liquid chromatography/quadrupole-Orbitrap mass spectrometry. J. Chromatogr. A 2019, 1584, 87–96. [Google Scholar] [CrossRef]
- Qiu, S.; Yang, W.Z.; Yao, C.L.; Qiu, Z.D.; Shi, X.J.; Zhang, J.X.; Hou, J.J.; Wang, Q.R.; Wu, W.Y.; Guo, D.A. Nontargeted metabolomic analysis and “commercial-homophyletic” comparison-induced biomarkers verification for the systematic chemical differentiation of five different parts of Panax ginseng. J. Chromatogr. A 2016, 1453, 78–87. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.H.; Kim, U.; Liu, J.B.; Cheng, C.J.; Wu, W.B.; Guo, S.; Feng, Y.J.; Quinn, R.J.; Hou, Y.Y.; Bai, G. Comprehensive TCM molecular networking based on MS/MS in silico spectra with integration of virtual screening and affinity MS screening for discovering functional ligands from natural herbs. Anal. Bioanal. Chem. 2019, 411, 5785–5797. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Z.; Ye, M.; Qiao, X.; Liu, C.F.; Miao, W.J.; Bo, T.; Tao, H.Y.; Guo, D.A. A strategy for efficient discovery of new natural compounds by integrating orthogonal column chromatography and liquid chromatography/mass spectrometry analysis: Its application in Panax ginseng, Panax quinquefolium and Panax notoginseng to characterize 437 potential new ginsenosides. Anal. Chim. Acta 2012, 739, 56–66. [Google Scholar] [CrossRef] [PubMed]
- Torres-Benitez, A.; Rivera-Montalvo, M.; Sepulveda, B.; Castro, O.N.; Nagles, E.; Simirgiotis, M.J.; Garcia-Beltran, O.; Areche, C. Metabolomic analysis of two Parmotrema Lichens: P-robustum (Degel.) Hale and P-andinum (Mull. Arg.) Hale using UHPLC-ESI-OT-MS-MS. Molecules 2017, 22, 1861. [Google Scholar] [CrossRef]
- Jia, L.; Zuo, T.T.; Zhang, C.X.; Li, W.W.; Wang, H.D.; Hu, Y.; Wang, X.Y.; Qian, Y.X.; Yang, W.Z.; Yu, H.S. Simultaneous profiling and holistic comparison of the metabolomes among the flower buds of Panax ginseng, Panax quinquefolius, and Panax notoginseng by UHPLC/IM-QTOF-HDMSE-based metabolomics analysis. Molecules 2019, 24, 2188. [Google Scholar] [CrossRef] [PubMed]
- May, J.C.; Morris, C.B.; McLean, J.A. Ion mobility collision cross section compendium. Anal. Chem. 2017, 89, 1032–1044. [Google Scholar] [CrossRef] [PubMed]
- Zhang, C.X.; Zuo, T.T.; Wang, X.Y.; Wang, H.D.; Hu, Y.; Li, Z.; Li, W.W.; Jia, L.; Qian, Y.X.; Yang, W.Z.; et al. Integration of data-dependent acquisition (DDA) and data-independent high-definition MSE (HDMSE) for the comprehensive profiling and characterization of multicomponents from Panax japonicus by UHPLC/IM-QTOF-MS. Molecules 2019, 24, 2708. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Q.; Xiao, X.H.; Li, M.; Li, W.H.; Yu, M.; Zhang, H.B.; Sun, X.F.; Mao, L.L.; Xiang, H.D. Attenuating effect of Fufang Xueshuantong Capsule on kidney function in diabetic nephropathy model. J. Nat. Med. 2013, 67, 86–97. [Google Scholar] [CrossRef] [PubMed]
- Duan, H.H.; Huang, J.M.; Li, W.; Tang, M.K. Protective effects of Fufang Xueshuantong on diabetic retinopathy in rats. Evid.-Based. Complement. Altern. Med. 2013, 408268. [Google Scholar] [CrossRef]
- Liu, H.; Zhang, W.J.; Long, C.F.; Su, W.W. Protective effects of traditional Chinese herbal formula Compound Xueshuantong Capsule (CXC) on rats with blood circulation disorders. Biotechnol. Biotechnol. Equip. 2017, 31, 846–854. [Google Scholar] [CrossRef]
- Sheng, S.J.; Wang, J.X.; Wang, L.R.; Liu, H.; Li, P.B.; Liu, M.H.; Long, C.F.; Xie, C.S.; Xie, X.Q.; Su, W.W. Network pharmacology analyses of the antithrombotic pharmacological mechanism of Fufang Xueshuantong Capsule with experimental support using disseminated intravascular coagulation rats. J. Ethnopharmacol. 2014, 154, 736–744. [Google Scholar] [CrossRef] [PubMed]
- Kang, J.; Zhou, L.; Li, Z.L.; Sun, Z.; Wang, X.H.; Wang, W.; Zuo, L.H.; Zhang, J.; Liu, L.W.; Shi, Y.Y.; et al. Chemical constituents study of compound Xueshuantong capsules based on UHPLC-Q-Orbitrap HRMS. Chin. J. Pharm. Anal. 2019, 39, 791–804. [Google Scholar]
- Zhou, C.H.; Su, L.F.; Shang, W.D.; Rong, Z.H.; Sun, M.M.; Zhang, K.; Shi, X.W.; Wang, Q. Simultaneous determination of eight chemicals in Fufang Xueshuantong Capsules by LC–MS-MS with periodic polarity switching. J. Chromatogr. Sci. 2015, 53, 1757–1764. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Guo, R.X.; Zhou, G.H.; Zhou, X.D.; Kou, Z.Z.; Sui, F.; Li, C.; Tang, L.Y.; Wang, Z.J. Traditional uses, botany, phytochemistry, pharmacology and toxicology of Panax notoginseng (Burk.) F.H. Chen: A review. J. Ethnopharmacol. 2014, 188, 234–258. [Google Scholar] [CrossRef]
- Yang, W.Z.; Hu, Y.; Wu, W.Y.; Ye, M.; Guo, D.A. Saponins in the genus Panax L. (Araliaceae): A systematic review of their chemical diversity. Phytochemistry 2014, 106, 7–24. [Google Scholar] [CrossRef] [PubMed]
- Lai, C.J.S.; Tan, T.; Zeng, S.L.; Qi, L.W.; Liu, X.G.; Dong, X.; Li, P.; Liu, E.H. An integrated high resolution mass spectrometric data acquisition method for rapid screening of saponins in Panax notoginseng (Sanqi). J. Pharm. Biomed. Anal. 2015, 109, 184–191. [Google Scholar] [CrossRef]
- Yang, W.Z.; Qiao, X.; Li, K.; Fan, J.R.; Bo, T.; Guo, D.A.; Ye, M. Identification and differentiation of Panax ginseng, Panax quinquefolium, and Panax notoginseng by monitoring multiple diagnostic chemical markers. Acta Pharm. Sin. B 2016, 6, 568–575. [Google Scholar] [CrossRef] [Green Version]
- Pang, H.Q.; Wu, L.; Tang, Y.P.; Zhou, G.S.; Qu, C.; Duan, J.A. Chemical analysis of the herbal medicine Salviae multiorrhizae Radix et Rhizoma (Danshen). Molecules 2016, 21, 51. [Google Scholar] [CrossRef]
- Guo, Z.Z.; Lou, Y.M.; Kong, M.Y.; Luo, Q.; Liu, Z.Q.; Wu, J.J. A systematic review of phytochemistry, pharmacology and pharmacokinetics on Astragali Radix: Implications for Astragali Radix as a personalized medicine. Int. J. Mol. Sci. 2019, 20, 1463. [Google Scholar] [CrossRef]
- Chen, Y.Y.; Zhai, S.; Xu, L.L.; Jiang, L.J.; Jiang, Y.Y.; Liu, B. Identification on chemical constituents of aqueous extract of Scrophulariae Radix by UPLC-LTQ-Orbitrap HRMS combined with cleavage pathways. Chin. Tradit. Herbal Drugs 2019, 50, 2822–2829. [Google Scholar]
- Qiu, S.; Yang, W.Z.; Shi, X.J.; Yao, C.L.; Yang, M.; Liu, X.; Jiang, B.H.; Wu, W.Y.; Guo, D.A. A green protocol for efficient discovery of novel natural compounds: Characterization of new ginsenosides from the stem and leaves of Panax ginseng. Anal. Chim. Acta 2015, 893, 65–76. [Google Scholar] [CrossRef] [PubMed]
- Yang, W.Z.; Bo, T.; Ji, S.; Qiao, X.; Guo, D.A.; Ye, M. Rapid chemical profiling of saponins in the flower buds of Panax notoginseng by integrating MCI gel column chromatography and liquid chromatography/mass spectrometry analysis. Food Chem. 2013, 139, 762–769. [Google Scholar] [CrossRef] [PubMed]
- Qiu, S.; Yang, W.Z.; Yao, C.L.; Shi, X.J.; Li, J.Y.; Lou, Y.; Duan, Y.N.; Wu, W.Y.; Guo, D.A. Malonylginsenosides with potential antidiabetic activities from the flower buds of Panax ginseng. J. Nat. Prod. 2017, 80, 899–908. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.J.; Yang, W.Z.; Qiu, S.; Yao, C.L.; Shen, Y.; Pan, H.Q.; Bi, Q.R.; Yang, M.; Wu, W.Y.; Guo, D.A. An in-source multiple collision-neutral loss filtering based nontargeted metabolomics approach for the comprehensive analysis of malonyl-ginsenosides from Panax ginseng, P quinquefolius, and P. notoginseng. Anal. Chim. Acta 2017, 952, 59–70. [Google Scholar] [CrossRef] [PubMed]
- Li, H.F.; Xu, F.; Yang, P.; Liu, G.X.; Shang, M.Y.; Wang, X.; Yin, J.; Cao, S.Q. Systematic screening and characterization of prototype constituents and metabolites of total astragalosides using HPLC-ESI-IT-TOF-MSn after oral administration to rats. J. Pharm. Biomed. Anal. 2017, 142, 102–112. [Google Scholar] [CrossRef] [PubMed]
- Yu, H.; Hu, Q.; Sun, J.; Feng, R.; Zhang, S.; Zhang, J.X.; Mao, X.H.; Ji, S. Comprehensive screening of multi-components in Huangqi Injection by ultra-performance liquid chromatography coupled with quadrupole-time-of-flight mass spectrometry. World Chin. Med. 2019, 14, 809–817. [Google Scholar]
- Yan, X.J.; Zheng, B.W.; Zhang, Y.D.; Li, N.A.; Liu, C.M.; Cao, Z.; Zhang, Q. Changes of chemical components in raw products and characteristic processed products with porcine cardiac blood of Salviae Miltiorrhizae Radix et Rhizoma from Menghe Medical School by UPLC-Q-TOF/MS. Chin. J. Exp. Tradit. Med. Formulae 2019, 25, 109–116. [Google Scholar]
- Wang, J.Z.; Xu, F.; Liu, Z.; Ma, L.M.; Shang, M.Y.; Liu, G.X.; Cai, S.Q. Identification of chemical constituents in Scrophulariae Radix by HPLC-IT-TOF-MS. Chin. J. Chin. Mater. Med. 2016, 41, 1257–1268. [Google Scholar] [CrossRef]
- Paglia, G.; Angel, P.; Williams, J.P.; Richardson, K.; Olivos, H.J.; Thompson, J.W.; Menikarachchi, L.; Lai, S.; Walsh, C.; Moseley, A.; et al. Ion mobility-derived collision cross section as an additional measure for lipid fingerprinting and identification. Anal. Chem. 2015, 87, 1137–1144. [Google Scholar] [CrossRef]
Sample Availability: Sample of Compound Xueshuantong Capsule is available from the authors. |







© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zuo, T.; Qian, Y.; Zhang, C.; Wei, Y.; Wang, X.; Wang, H.; Hu, Y.; Li, W.; Wu, X.; Yang, W. Data-Dependent Acquisition and Database-Driven Efficient Peak Annotation for the Comprehensive Profiling and Characterization of the Multicomponents from Compound Xueshuantong Capsule by UHPLC/IM-QTOF-MS. Molecules 2019, 24, 3431. https://doi.org/10.3390/molecules24193431
Zuo T, Qian Y, Zhang C, Wei Y, Wang X, Wang H, Hu Y, Li W, Wu X, Yang W. Data-Dependent Acquisition and Database-Driven Efficient Peak Annotation for the Comprehensive Profiling and Characterization of the Multicomponents from Compound Xueshuantong Capsule by UHPLC/IM-QTOF-MS. Molecules. 2019; 24(19):3431. https://doi.org/10.3390/molecules24193431
Chicago/Turabian StyleZuo, Tiantian, Yuexin Qian, Chunxia Zhang, Yuxi Wei, Xiaoyan Wang, Hongda Wang, Ying Hu, Weiwei Li, Xiaohui Wu, and Wenzhi Yang. 2019. "Data-Dependent Acquisition and Database-Driven Efficient Peak Annotation for the Comprehensive Profiling and Characterization of the Multicomponents from Compound Xueshuantong Capsule by UHPLC/IM-QTOF-MS" Molecules 24, no. 19: 3431. https://doi.org/10.3390/molecules24193431
APA StyleZuo, T., Qian, Y., Zhang, C., Wei, Y., Wang, X., Wang, H., Hu, Y., Li, W., Wu, X., & Yang, W. (2019). Data-Dependent Acquisition and Database-Driven Efficient Peak Annotation for the Comprehensive Profiling and Characterization of the Multicomponents from Compound Xueshuantong Capsule by UHPLC/IM-QTOF-MS. Molecules, 24(19), 3431. https://doi.org/10.3390/molecules24193431