New Insights into the Reaction Paths of Hydroxyl Radicals with Purine Moieties in DNA and Double-Stranded Oligodeoxynucleotides
Abstract
:1. Introduction
2. Results and Discussion
2.1. Radiolytic Production of Transients
2.2. Pulse Radiolysis in Aqueous Solutions
2.2.1. Comparison of PMT and ICCD Detection Methods Using Nucleosides
2.2.2. Mixture of dC and dG Nucleosides
2.2.3. Mixture of dC, dG, T and dA Nucleosides vs. Calf-Thymus DNA (ct-DNA)
2.2.4. Single Stranded 12-Mer Oligodeoxynucleotides
2.2.5. Double-Stranded 12-Mer Oligodeoxynucleotides
2.3. γ-Radiolysis
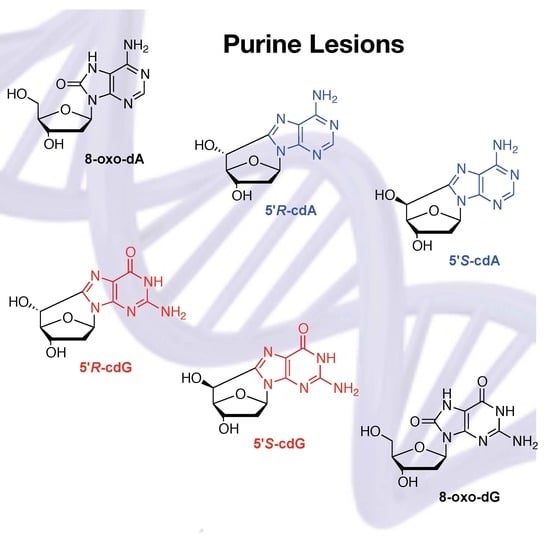
2.3.1. Hydroxyl Radical–Induced Formation of Purine Lesions in ct-DNA
- (i)
- the commercial ct-DNA solution containing 1 mM Tris-HCl, pH 7.5, with 1 mM NaCl and 1 mM EDTA was firstly lyophilized and then 200 µL of a N2O saturated ct-DNA aqueous solutions (0.5 mg/mL) at natural pH were prepared;
- (ii)
- the commercial ct-DNA solution was desalted by ethanol precipitation (removal of the additives Tris-HCl, NaCl and EDTA) and then 200 µL of a N2O saturated ct-DNA (0.5 mg/mL) were prepared in 50 mM phosphate buffer, pH 7.2
2.3.2. Hydroxyl Radical–Induced Formation of Purine Lesions in Double Stranded 21-Mer Oligonucleotides
2.4. Hydroxyl Radical Generated by Fenton Reactions and Formation of Purine Lesions in Double Stranded 21-Mer Oligonucleotides
3. Materials and Methods
3.1. Chemicals, Reagents and Enzymes
3.2. Oligodeoxynucleotides (ODNs) Synthesis and Purification
3.3. Preparation of Double Stranded Oligonucleotide Substrates
3.4. Pulse Radiolysis
3.5. γ-Radiolysis Experiments
3.6. Fenton-Type Reagent Treatments of ds-ODNs
3.6.1. CuCl2 with L-Methionine
3.6.2. Kinetic Study by Cu2+/H2O2 of ds-ODNs with L-Methionine
3.6.3. Kinetic Study by Fe2+/H2O2) of ds-ODNs with L-Methionine
3.7. Enzymatic Digestion of the ct-DNA and ds-ODNs
3.8. HPLC Analysis and Quantification of Modified Nucleosides by Stable Isotope LC-MS/MS
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Greenberg, M.M. (Ed.) Radical and Radical Ion Reactivity in Nucleic Acid Chemistry; John Wiley & Sons: Honoken, NJ, USA, 2009. [Google Scholar]
- von Sonntag, C. Free-Radical-Induced DNA Damage and Its Repair. A Chemical Perspective; Springer Science: Berlin/Heidelberg, Germany, 2006. [Google Scholar]
- Chatgilialoglu, C.; Ferreri, C.; Terzidis, M.A. Purine 5′,8-cyclonucleoside lesions: Chemistry and biology. Chem. Soc. Rev. 2011, 40, 1368–1382. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; Ferreri, C.; Geacintov, N.E.; Krokidis, M.G.; Liu, Y.; Masi, A.; Shafirovich, V.; Terzidis, M.A.; Tsegay, P.S. 5′,8-Cyclopurine Lesions in DNA Damage: Chemical, Analytical, Biological and Diagnostic Significance. Cells 2019, 8, 513. [Google Scholar] [CrossRef] [PubMed]
- Adhikary, A.; Becker, D.; Palmer, B.J.; Heizer, A.N.; Sevilla, M.D. Direct formation of the C5′-radical in the sugar-phosphate backbone of DNA by high energy radiation. J. Phys. Chem. B 2012, 116, 5900–5906. [Google Scholar] [CrossRef] [PubMed]
- Mitra, D.; Luo, X.; Morgan, A.; Wang, J.; Hoang, M.P.; Lo, J.; Guerrero, C.R.; Lennerz, J.K.; Mihm, M.C.; Wargo, J.A.; et al. An ultraviolet-radiation-independent pathway to melanoma carcinogenesis in the red hair/fair skin background. Nature 2012, 491, 449–453. [Google Scholar] [CrossRef] [PubMed]
- Wang, J.; Clauson, C.L.; Robbins, P.D.; Niedernhofer, L.J.; Wang, Y. The oxidative DNA lesions 8,5′-cyclopurines accumulate with aging in a tissue-specific manner. Aging Cell 2012, 11, 714–716. [Google Scholar] [CrossRef] [PubMed]
- Yu, Y.; Guerrero, C.R.; Liu, S.; Amato, N.J.; Sharma, Y.; Gupta, S.; Wang, Y. Comprehensive assessment of oxidatively induced modifications of DNA in a rat model of human Wilson’s disease. Mol. Cell. Proteom. 2016, 15, 810–817. [Google Scholar] [CrossRef] [PubMed]
- Robinson, A.R.; Yousefzadeh, M.J.; Rozgaja, T.A.; Wang, J.; Li, X.; Tilstra, J.S.; Feldman, C.H.; Gregg, S.Q.; Johnson, C.H.; Skoda, E.M.; et al. Spontaneous DNA damage to the nuclear genome promotes senescence, redox imbalance and aging. Redox Biol. 2018, 17, 259–273. [Google Scholar] [CrossRef]
- Krokidis, M.; Louka, M.; Efthimiadou, E.; Zervou, S.-K.; Papadopoulos, K.; Hiskia, A.; Ferreri, C.; Chatgilialoglu, C. Membrane lipidome reorganization and accumulation of tissue DNA lesions in tumor-bearing mice: An exploratory study. Cancers 2019, 11, 480. [Google Scholar] [CrossRef]
- Kropachev, K.; Ding, S.; Terzidis, M.A.; Masi, A.; Liu, Z.; Cai, Y.; Kolbanovskiy, M.; Chatgilialoglu, C.; Broyde, S.; Geancitov, N.E.; et al. Structural basis for the recognition of diastereomeric 5′,8-cyclo-2′-deoxypurine lesions by the human nucleotide excision repair system. Nucleic. Acids Res. 2014, 42, 5020–5032. [Google Scholar] [CrossRef]
- Shafirovich, V.; Kolbanovskiy, M.; Kropachev, K.; Liu, Z.; Cai, Y.; Terzidis, M.A.; Masi, A.; Chatgilialoglu, C.; Amin, S.; Dadali, A.; et al. Nucleotide excision repair and impact of site-specific 5′,8-cyclopurine and bulky DNA lesions on the physical properties of nucleosomes. Biochemistry 2019, 58, 561–574. [Google Scholar] [CrossRef]
- Yu, Y.; Wang, P.; Cui, Y.; Wang, Y. Chemical analysis of DNA damage. Anal. Chem. 2018, 90, 556–576. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.; Ye, W.; Prestwich, E.G.; Wishnok, J.S.; Taghizadeh, K.; Dedon, P.C.; Tannenbaum, S.R. Comparative analysis of four oxidized guanine lesions from reactions of DNA with peroxynitrite, single oxygen, and γ-radiation. Chem Res. Toxicol. 2013, 26, 195–202. [Google Scholar] [CrossRef] [PubMed]
- Dizdaroglu, M.; Coskun, E.; Jaruga, P. Measurement of oxidatively induced DNA damage and its repair, by mass spectrometric techniques. Free Radic. Res. 2015, 49, 525–548. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C. Cuclopurine (cPu) lesions: What, how and why. Free Radic. Res. 2019, 53, 941–943. [Google Scholar] [CrossRef]
- Baldacchino, G.; Hickel, B.A. method to improve the nonrepetitive acquisition of transient absorption spectra with an intensified charge-coupled device camera. Rev. Sci. Instrum. 1998, 69, 1605–1609. [Google Scholar] [CrossRef]
- Nauser, T.; Jacoby, M.; Koppenol, W.H.; Squier, T.C.; Schöneich, C. Calmoduline methionine residues are targets for one electron oxidation by hydroxyl radicals: Formation of S∴N three-electron bonded radical complexes. Chem. Commun. 2005, 587–589. [Google Scholar] [CrossRef]
- Buxton, G.V.; Greenstock, C.L.; Helman, W.P.; Ross, A.B. Critical review of rate constants for hydrated electrons, hydrogen atoms and hydroxyl radicals (OH/O−) in aqueous solution. J. Phys. Chem. Ref. Data 1988, 17, 513–886. [Google Scholar] [CrossRef]
- Ross, A.B.; Mallard, W.G.; Helman, W.P.; Buxton, G.V.; Huie, R.E.; Neta, P. NDRLNIST Solution Kinetic Database-Ver. 3; Notre Dame Radiation Laboratory, Notre Dame, IN and NIST Standard Reference Data: Gaithersburg, MD, USA, 1998. [Google Scholar]
- Hissung, A.; von Sonntag, C. Radiolysis of Cytosine, 5-Methyl Cytosine and 2′-Deoxycytidine in Deoxygenated Aqueous Solution. A Pulse Spectroscopic and Pulse Conductometric Study of the OH Adduct. Z. Nat. 1978, 33, 321–328. [Google Scholar] [CrossRef]
- Aravandikumar, C.T.; Schuchmann, M.N.; Rao, B.S.M.; von Sonntag, J.; von Sonntag, C. The reactions of cytidine and 2′deoxycytidine with SO4•− revisited. Pulse radiolysis and product studies. Org. Biomol. Chem. 2003, 1, 401–408. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; D’Angelantonio, M.; Guerra, M.; Kaloudis, P.; Mulazzani, Q.G. A reevaluation of the ambident reactivity of guanine moiety towards hydroxyl radicals. Angew. Chem. Int. Ed. 2009, 48, 2214–2217. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Caminal, C.; Guerra, M.; Mulazzani, Q.G. Tautomers of one-electron-oxidized guanosine. Angew. Chem. Int. Ed. 2005, 44, 6030–6032. [Google Scholar] [CrossRef] [PubMed]
- Chatgilialoglu, C.; D’Angelantonio, M.; Kciuk, G.; Bobrowski, K. New Insights into the Reaction Paths of Hydroxyl Radicals with 2′-Deoxyguanosine. Chem. Res. Toxicol. 2011, 24, 2200–2206. [Google Scholar] [CrossRef] [PubMed]
- Vieira, A.J.S.C.; Steenken, S. Pattern of OH Radical Reaction with Adenine and Its Nucleosides and Nucleotides. Characterization of Two Types of Isomeric OH Adduct and Their Unimolecular Transformation Reactions. J. Am. Chem. Soc. 1990, 112, 6986–6994. [Google Scholar] [CrossRef]
- Wu, L.D.; Liu, K.H.; Jie, J.L.; Song, D.; Su, H.M. Direct Observation of Guanine Radical Cation Deprotonation in G-Quadruplex DNA. J. Am. Chem. Soc. 2015, 137, 259–266. [Google Scholar] [CrossRef] [PubMed]
- Banyasz, A.; Martinez-Fernandez, L.; Balty, C.; Perron, M.; Douki, T.; Improta, R.; Markovitsi, D. Absorption of Low-Energy UV Radiation by Human Telomere G-Quadruplexes Generates Long-Lived Guanine Radical Cations. J. Am. Chem. Soc. 2017, 139, 10561–10568. [Google Scholar] [CrossRef]
- Balanikas, E.; Banyasz, A.; Baldacchino, G.; Markovitsi, D. Populations and Dynamics of Guanine Radicals in DNA strands—Direct versus Indirect Generation. Molecules 2019, 24, 2347. [Google Scholar] [CrossRef]
- Capobianco, A.; Caruso, T.; D’Urci, A.M.; Fusco, S.; Masi, A.; Scrima, M.; Chatgilialoglu, C.; Peluso, A. Delocalized hole domains in guanine rich DNA oligonucleotides. J. Phys. Chem. B 2015, 119, 5462–5466. [Google Scholar] [CrossRef]
- Terzidis, M.A.; Chatgilialoglu, C. An ameliorative protocol for the quantification of purine 5′,8-cyclo-2′-deoxynucleosides in oxidized DNA. Front. Chem. 2015, 3, 47. [Google Scholar] [CrossRef]
- Krokidis, M.G.; Terzidis, M.A.; Efthimiadou, E.; Zervou, S.K.; Kordas, G.; Papadopoulos, K.; Hiskia, A.; Kletsas, D.; Chatgilialoglu, C. Purine 5′,8-cyclo-2′-deoxynucleoside lesions: Formation by radical stress and repair in human breast epithelial cancer cells. Free Radic. Res. 2017, 51, 470–482. [Google Scholar] [CrossRef]
- Terzidis, M.A.; Ferreri, C.; Chatgilialoglu, C. Radiation-induced formation of purine lesions in single and double stranded DNA: Revised quantification. Front. Chem. 2015, 3, 18. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Bazzanini, R.; Jimenez, L.B.; Miranda, M.A. (5′S)- and (5′R)-5′,8-cyclo-2′-deoxyguanosine: Mechanistic insights on the 2′-deoxyguanosin-5′-yl radical cyclization. Chem. Res. Toxicol. 2007, 20, 1820–1824. [Google Scholar] [CrossRef] [PubMed]
- Boussicault, F.; Kaloudis, P.; Caminal, C.; Mulazzani, Q.G.; Chatgilialoglu, C. The fate of C5′ radicals of purine nucleosides under oxidative conditions. J. Am. Chem. Soc. 2008, 130, 8377–8385. [Google Scholar] [CrossRef] [PubMed]
- Belmadoui, N.; Boussicault, F.; Guerra, M.; Ravanat, J.-L.; Chatgilialoglu, C.; Cadet, J. Radiation-induced formation of purine 5′,8-cyclonucleosides in isolated and cellular DNA: High stereospecificity and modulating effect of oxygen. Org. Biomol. Chem. 2010, 8, 3211–3219. [Google Scholar] [CrossRef] [PubMed]
- Terzidis, M.A.; Prisecaru, A.; Molphy, Z.; Barron, N.; Randazzo, A.; Dumont, E.; Krokidis, M.G.; Kellett, A.; Chatgilialoglu, C. Radical-induced purine lesion formation is dependent on DNA helical topology. Free Radic. Res. 2016, 50, S91–S101. [Google Scholar] [CrossRef] [Green Version]
- Miller, D.M.; Buettner, G.R.; Aust, S.D. Transition metals as catalysts of “autoxidation” reactions. Free Radic. Biol. Med. 1990, 8, 95–108. [Google Scholar] [CrossRef]
- Stohs, S.J.; Bagchi, D. Oxidative mechanisms in the toxicity of metal ions. Free Radic. Biol. Med. 1995, 18, 321–336. [Google Scholar] [CrossRef] [Green Version]
- Haliwell, B.; Gutteridge, J.M.C. Free Radicals in Biology and Medicine, 5th ed.; Oxford University Press: Oxford, UK, 2015. [Google Scholar]
- Guerrero, C.R.; Wang, J.; Wang, Y. Induction of 8,5′-cyclo-2′-deoxyadenosine and 8,5′-cyclo-2′-deoxyguanosine in isolated DNA by Fenton-type reagents. Chem. Res. Toxicol. 2013, 26, 1361–1366. [Google Scholar] [CrossRef]
- Bergeron, F.; Auvré, F.; Radicella, J.P.; Ravanat, J.-L. HO• radicals induce an unexpected high proportion of tandem base lesions refractory to repair by DNA glycosylases. Proc. Natl. Acad. Sci. USA 2010, 107, 5528–5533. [Google Scholar] [CrossRef]
- Flyunt, R.; Bazzanini, R.; Chatgilialoglu, C.; Mulazzani, Q.G. Fate of the 2′-Deoxyadenosin-5′-yl Radical under Anaerobic ConditionsJ. Am. Chem. Soc. 2000, 122, 4225–4226. [Google Scholar] [CrossRef]
- Chatgilialoglu, C.; Guerra, M.; Mulazzani, Q.G. Model studies of DNA C5′ radicals. Selective generation and reactivity of 2′-deoxyadenosin-5’-yl radical. J. Am. Chem. Soc. 2003, 125, 3839–3848. [Google Scholar] [CrossRef]
Sample Availability: Samples of the compounds are not available from the authors. |















| Strands | Sequence (5′-3′) | Length |
|---|---|---|
| ODN1 | CGT ATG GTA TCG | 12 |
| ODN2 | CGA TAC CAT ACG | 12 |
| ODN3 | CGA TGG GGT ACG | 12 |
| ODN4 | CGT ACC CCA TCG | 12 |
| ODN5 | GGG (TTA GGG)3 | 21 |
| ODN6 | CCC (TAA CCC)3 | 21 |
| Lesion | Lesions/107 nu/Gy a | 5′R/5′S a | Lesions/107 nu/Gy b | 5′R/5′S b | Lesions/107 nu/Gy c | 5′R/5′S c |
|---|---|---|---|---|---|---|
| 8-oxo-dG | 171.8 ± 13.0 | 200.1 ± 3.03 | 780 | |||
| 8-oxo-dA | 22.22 ± 1.25 | 28.04 ± 0.46 | 72 | |||
| 5′R-cdG | 1.40 ± 0.12 | 4.5 | 2.98 ± 0.10 | 4.7 | 151 d | ~3 |
| 5′S-cdG | 0.31 ± 0.02 | 0.64 ± 0.06 | 50 d | |||
| 5′R-cdA | 1.55 ± 0.08 | 1.2 | 1.47 ± 0.14 | 1.5 | 114d | ~4 |
| 5′S-cdA | 1.30 ± 0.06 | 0.95 ± 0.07 | 28d |
| ds-(ODN5/ODN6) | ||
|---|---|---|
| Lesion | Lesions/107 dG/Gy | Lesions/107 dA/Gy |
| 8-oxo-dG | 13.49 ± 1.87 | |
| 8-oxo-dA | 2.11 ± 0.30 | |
| 5′R-cdG | 0.32 ± 0.04 | |
| 5′S-cdG | 0.67 ± 0.18 | |
| 5′R-cdA | 1.60 ± 0.29 | |
| 5′S-cdA | 1.38 ± 0.27 | |
| Lesions | From Cu1+ | From Fe2+ |
|---|---|---|
| 5′R-cdG | 0.16 | 0.18 |
| 5′S-cdG | 0.17 | 0.21 |
| 5′R-cdA | 0.20 | 0.21 |
| 5′S-cdA | 0.14 | 0.14 |
| 8-oxo-dG | 3.88 | 4.81 |
| 8-oxo-dA | 0.60 | 0.49 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Chatgilialoglu, C.; Krokidis, M.G.; Masi, A.; Barata-Vallejo, S.; Ferreri, C.; Terzidis, M.A.; Szreder, T.; Bobrowski, K. New Insights into the Reaction Paths of Hydroxyl Radicals with Purine Moieties in DNA and Double-Stranded Oligodeoxynucleotides. Molecules 2019, 24, 3860. https://doi.org/10.3390/molecules24213860
Chatgilialoglu C, Krokidis MG, Masi A, Barata-Vallejo S, Ferreri C, Terzidis MA, Szreder T, Bobrowski K. New Insights into the Reaction Paths of Hydroxyl Radicals with Purine Moieties in DNA and Double-Stranded Oligodeoxynucleotides. Molecules. 2019; 24(21):3860. https://doi.org/10.3390/molecules24213860
Chicago/Turabian StyleChatgilialoglu, Chryssostomos, Marios G. Krokidis, Annalisa Masi, Sebastian Barata-Vallejo, Carla Ferreri, Michael A. Terzidis, Tomasz Szreder, and Krzysztof Bobrowski. 2019. "New Insights into the Reaction Paths of Hydroxyl Radicals with Purine Moieties in DNA and Double-Stranded Oligodeoxynucleotides" Molecules 24, no. 21: 3860. https://doi.org/10.3390/molecules24213860
APA StyleChatgilialoglu, C., Krokidis, M. G., Masi, A., Barata-Vallejo, S., Ferreri, C., Terzidis, M. A., Szreder, T., & Bobrowski, K. (2019). New Insights into the Reaction Paths of Hydroxyl Radicals with Purine Moieties in DNA and Double-Stranded Oligodeoxynucleotides. Molecules, 24(21), 3860. https://doi.org/10.3390/molecules24213860