Geographical Authentication of Macrohyporia cocos by a Data Fusion Method Combining Ultra-Fast Liquid Chromatography and Fourier Transform Infrared Spectroscopy
Abstract
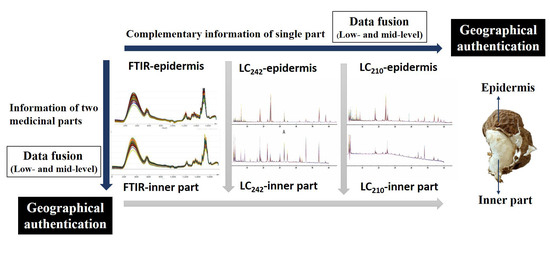
:1. Introduction
2. Results and Discussion
2.1. Spectral Analysis
2.2. Quantitative Analysis of Five Triterpene Acids
2.3. Chromatographic Data Preprocessing
2.4. PLS-DA Using Chromatograms and FTIR Spectra
2.5. Low-Level Data Fusion
2.5.1. PLS-DA of Poria
2.5.2. PLS-DA of Poriae Cutis
2.5.3. PLS-DA of Combination Data of Two Medicinal Parts
2.6. Mid-Level Data Fusion
2.6.1. The Extraction of Feature Variables
2.6.2. PLS-DA of Poria
2.6.3. PLS-DA of Poriae Cutis
2.6.4. PLS-DA of Combination Data of Two Medicinal Parts
3. Materials and Methods
3.1. Reagents, Solvents and Standard References
3.2. Samples
3.3. FTIR Spectra Acquisition
3.4. Chromatographic Analysis
3.5. Method Validation
3.6. Preprocessing of Chromatograms and Spectra
3.7. Multiple Chromatograms and Spectra Data Fusion
3.8. Evaluation of Model Performance
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Khan, I.; Huang, G.; Li, X.; Leong, W.; Xia, W.; Hsiao, W.L.W. Mushroom polysaccharides from Ganoderma lucidum and Poria cocos reveal prebiotic functions. J. Funct. Foods 2018, 41, 191–201. [Google Scholar] [CrossRef]
- Miao, H.; Zhao, Y.; Vaziri, N.D.; Tang, D.; Chen, H.; Chen, H.; Khazaeli, M.; Tarbiat-Boldaji, M.; Hatami, L.; Zhao, Y. Lipidomics biomarkers of diet-induced hyperlipidemia and its treatment with Poria cocos. J. Agric. Food Chem. 2016, 64, 969–979. [Google Scholar] [CrossRef]
- Lee, S.; Lee, S.; Roh, H.; Song, S.; Ryoo, R.; Pang, C.; Baek, K.; Kim, K. Cytotoxic constituents from the sclerotia of Poria cocos against human lung adenocarcinoma cells by inducing mitochondrial apoptosis. Cells 2018, 7, 116. [Google Scholar] [CrossRef]
- Wu, K.; Fan, J.; Huang, X.; Wu, X.; Guo, C. Hepatoprotective effects exerted by Poria cocos polysaccharides against acetaminophen-induced liver injury in mice. Int. J. Biol. Macromol. 2018, 114, 137–142. [Google Scholar] [CrossRef] [PubMed]
- Lee, S.; Choi, E.; Yang, S.; Ryoo, R.; Moon, E.; Kim, S.; Kim, K.H. Bioactive compounds from sclerotia extract of Poria cocos that control adipocyte and osteoblast differentiation. Bioorg. Chem. 2018, 81, 27–34. [Google Scholar] [CrossRef] [PubMed]
- Zhu, L.; Xu, J.; Zhang, S.; Wang, R.; Huang, Q.; Chen, H.; Dong, X.; Zhao, Z. Qualitatively and quantitatively comparing secondary metabolites in three medicinal parts derived from Poria cocos (Schw.) Wolf using UHPLC-QTOF-MS/MS-based chemical profiling. J. Pharm. Biomed. 2018, 150, 278–286. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Jin, H.; Liu, H.; Wang, Y. Ultraviolet spectroscopy combined with ultra-fast liquid chromatography and multivariate statistical analysis for quality assessment of wild Wolfiporia extensa from different geographical origins. Spectrochim. Acta Part A 2016, 165, 61–68. [Google Scholar] [CrossRef]
- Biancolillo, A.; Marini, F. Chapter four—Chemometrics applied to plant spectral analysis. In Vibrational Spectroscopy for Plant Varieties and Cultivars Characterization, Comprehensive Analytical Chemistry, 1st ed.; Lopes, J., Sousa, C., Eds.; Elsevier: Amsterdam, The Netherlands, 2018; Volume 80, pp. 69–104. [Google Scholar]
- Yuan, T.; Zhao, Y.; Zhang, J.; Wang, Y. Application of variable selection in the origin discrimination of Wolfiporia cocos (F.A. Wolf) Ryvarden & Gilb. based on near infrared spectroscopy. Sci. Rep. 2018, 8, 89. [Google Scholar]
- Zhu, L.; Xu, J.; Wang, R.; Li, H.; Tan, Y.; Chen, H.; Dong, X.; Zhao, Z. Correlation between quality and geographical origins of Poria cocos revealed by qualitative fingerprint profiling and quantitative determination of triterpenoid acids. Molecules 2018, 23, 2200. [Google Scholar] [CrossRef]
- Chen, J.; Sun, S.; Ma, F.; Zhou, Q. Vibrational microspectroscopic identification of powdered traditional medicines: Chemical micromorphology of Poria observed by infrared and Raman microspectroscopy. Spectrochim. Acta Part A 2014, 128, 629–637. [Google Scholar] [CrossRef]
- Orlandi, G.; Calvini, R.; Foca, G.; Pigani, L.; Vasile Simone, G.; Ulrici, A. Data fusion of electronic eye and electronic tongue signals to monitor grape ripening. Talanta 2019, 195, 181–189. [Google Scholar] [CrossRef]
- Wu, X.; Zhang, Q.; Wang, Y. Traceability of wild Paris polyphylla Smith var. yunnanensis based on data fusion strategy of FT-MIR and UV-Vis combined with SVM and random forest. Spectrochim. Acta Part A 2018, 205, 479–488. [Google Scholar] [CrossRef]
- Ni, Y.; Li, B.; Kokot, S. Discrimination of Radix Paeoniae varieties on the basis of their geographical origin by a novel method combining high-performance liquid chromatography and Fourier transform infrared spectroscopy measurements. Anal. Methods-UK 2012, 4, 4326. [Google Scholar] [CrossRef]
- Borràs, E.; Ferré, J.; Boqué, R.; Mestres, M.; Aceña, L.; Busto, O. Data fusion methodologies for food and beverage authentication and quality assessment—A review. Anal. Chim. Acta 2015, 891, 1–14. [Google Scholar] [CrossRef]
- Casale, M.; Bagnasco, L.; Zotti, M.; Di Piazza, S.; Sitta, N.; Oliveri, P. A NIR spectroscopy-based efficient approach to detect fraudulent additions within mixtures of dried porcini mushrooms. Talanta 2016, 160, 729–734. [Google Scholar] [CrossRef]
- Bureau, S.; Cozzolino, D.; Clark, C.J. Contributions of Fourier-transform mid infrared (FT-MIR) spectroscopy to the study of fruit and vegetables: A review. Postharvest. Biol. Technol. 2019, 148, 1–14. [Google Scholar] [CrossRef]
- Li, Y.; Zhang, J.; Wang, Y. FT-MIR and NIR spectral data fusion: A synergetic strategy for the geographical traceability of Panax notoginseng. Anal. Bioanal. Chem. 2018, 410, 91–103. [Google Scholar] [CrossRef]
- Wu, Z.; Zhao, Y.; Zhang, J.; Wang, Y. Quality assessment of Gentiana rigescens from different geographical origins using FT-IR spectroscopy combined with HPLC. Molecules 2017, 22, 1238. [Google Scholar] [CrossRef]
- Wang, Y.; Shen, T.; Zhang, J.; Huang, H.; Wang, Y. Geographical authentication of Gentiana rigescens by high-performance liquid chromatography and infrared spectroscopy. Anal. Lett. 2018, 51, 2173–2191. [Google Scholar] [CrossRef]
- Obisesan, K.A.; Jiménez-Carvelo, A.M.; Cuadros-Rodriguez, L.; Ruisánchez, I.; Callao, M.P. HPLC-UV and HPLC-CAD chromatographic data fusion for the authentication of the geographical origin of palm oil. Talanta 2017, 170, 413–418. [Google Scholar] [CrossRef]
- Lucio-Gutiérrez, J.R.; Garza-Juárez, A.; Coello, J.; Maspoch, S.; Salazar-Cavazos, M.L.; Salazar-Aranda, R.; Waksman De Torres, N. Multi-wavelength high-performance liquid chromatographic fingerprints and chemometrics to predict the antioxidant activity of Turnera diffusa as part of its quality control. J. Chromatogr. A 2012, 1235, 68–76. [Google Scholar] [CrossRef] [PubMed]
- Lucio-Gutiérrez, J.R.; Coello, J.; Maspoch, S. Enhanced chromatographic fingerprinting of herb materials by multi-wavelength selection and chemometrics. Anal. Chim. Acta 2012, 710, 40–49. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Liu, Y.; Liu, Z.; Wang, C.; Song, Z.; Liu, Y.; Dong, Y.; Ning, Z.; Lu, A. Comparison of the roots of Salvia miltiorrhiza Bunge (Danshen) and its variety S. miltiorrhiza Bge f. Alba (Baihua Danshen) based on multi-wavelength HPLC-fingerprinting and contents of nine active components. Anal. Methods-UK 2016, 8, 3171–3182. [Google Scholar] [CrossRef]
- Horn, B.; Esslinger, S.; Pfister, M.; Fauhl-Hassek, C.; Riedl, J. Non-targeted detection of paprika adulteration using mid-infrared spectroscopy and one-class classification–Is it data preprocessing that makes the performance? Food Chem. 2018, 257, 112–119. [Google Scholar] [CrossRef]
- Cael, J.J.; Koenig, J.L.; Blackwell, J. Infrared and Raman spectroscopy of carbohydrates. Part VI: Normal coordinate analysis of V-amylose. Biopolymers 1975, 14, 1885–1903. [Google Scholar] [CrossRef]
- Li, S.; Wang, L.; Song, C.; Hu, X.; Sun, H.; Yang, Y.; Lei, Z.; Zhang, Z. Utilization of soybean curd residue for polysaccharides by Wolfiporia extensa (Peck) Ginns and the antioxidant activities in vitro. J. Taiwan Inst. Chem. E 2014, 45, 6–11. [Google Scholar] [CrossRef]
- Akihisa, T.; Uchiyama, E.; Kikuchi, T.; Tokuda, H.; Suzuki, T.; Kimura, Y. Anti-tumor-promoting effects of 25-methoxyporicoic acid A and other triterpene acids from Poria cocos. J. Nat. Prod. 2009, 72, 1786–1792. [Google Scholar] [CrossRef]
- Lee, S.; Lee, D.; Lee, S.O.; Ryu, J.; Choi, S.; Kang, K.S.; Kim, K.H. Anti-inflammatory activity of the sclerotia of edible fungus, Poria cocos Wolf and their active lanostane triterpenoids. J. Funct. Foods 2017, 32, 27–36. [Google Scholar] [CrossRef]
- Ying, Y.; Shan, W.; Zhang, L.; Zhan, Z. Lanostane triterpenes from Ceriporia lacerate HS-ZJUT-C13A, a fungal endophyte of Huperzia serrata. Helv. Chim. Acta 2013, 95, 2092–2097. [Google Scholar] [CrossRef]
- Maquelin, K.; Kirschner, C.; Choo-Smith, L.P.; van den Braak, N.; Endtz, H.P.; Naumann, D.; Puppels, G.J. Identification of medically relevant microorganisms by vibrational spectroscopy. J. Microbiol. Methods 2002, 51, 255–271. [Google Scholar] [CrossRef]
- Skov, T.; van den Berg, F.; Tomasi, G.; Bro, R. Automated alignment of chromatographic data. J. Chemom. 2006, 20, 484–497. [Google Scholar] [CrossRef]
- Ballabio, D.; Consonni, V. Classification tools in chemistry. Part 1: Linear models. PLS-DA. Anal. Methods-UK 2013, 5, 3790. [Google Scholar] [CrossRef]
- Ståhle, L.; Wold, S. Partial least squares analysis with cross-validation for the two-class problem: A Monte Carlo study. J. Chemom. 1987, 1, 185–196. [Google Scholar] [CrossRef]
- Barker, M.; Rayens, W. Partial least squares for discrimination. J. Chemom. 2003, 17, 166–173. [Google Scholar] [CrossRef]
- Sjöström, M.; Wold, S.; Söderström, B. PLS discriminant plots. In Pattern Recognition in Practice; Gelsema, E.S., Kanal, L.N., Eds.; Elsevier: Amsterdam, The Netherlands, 1986; pp. 461–470. [Google Scholar]
- Wold, S.; Johansson, E.; Cocchi, M. PLS: Partial least squares projections to latent structures. In 3D QSAR in Drug Design: Theory, Methods and Applications; Kubinyi, H., Ed.; KLUWER ESCOM Science Publisher: Leiden, The Netherlands, 1993; pp. 523–550. [Google Scholar]
- Wold, S.; Esbensen, K.; Geladi, P. Principal component analysis. Chemom. Intell. Lab. 1987, 2, 37–52. [Google Scholar] [CrossRef]
- Kennard, R.W.; Stone, L.A. Computer aided design of experiments. Technometrics 1969, 11, 137–148. [Google Scholar] [CrossRef]
- Oliveri, P.; Downey, G. Multivariate class modeling for the verification of food-authenticity claims. TrAC Trends Anal. Chem. 2012, 35, 74–86. [Google Scholar] [CrossRef]
- Aa, J.Y. Analysis of metabolomic data: Principal component analysis. Chin. J. Clin. Pharmacol. Ther. 2010, 15, 481–489. [Google Scholar]
Sample Availability: Samples of the compounds are not available from the authors. |







| Fusion Approach | Data Matrix | Calibration Set | Validation Set | |||
|---|---|---|---|---|---|---|
| R2(cum) | Q2(cum) | Accuracy | Accuracy | |||
| single technique | Poria | FTIR | 0.8883 | 0.7268 | 100% | 92.31% |
| LC242 | 0.6634 | 0.5277 | 96.15% | 100% | ||
| LC210 | 0.5174 | 0.4012 | 90.38% | 76.92% | ||
| Poria Cutis | FTIR | 0.9292 | 0.6981 | 100% | 96.15% | |
| LC242 | 0.2874 | 0.2204 | 65.38% | 34.62% | ||
| low-level data fusion | Poria | FTIR-LC242 | 0.9599 | 0.7917 | 100% | 100% |
| FTIR-LC210 | 0.9468 | 0.7663 | 100% | 100% | ||
| LC242-210 | 0.8097 | 0.6547 | 98.08% | 92.31% | ||
| FTIR-LC242-210 | 0.8823 | 0.7566 | 100% | 100% | ||
| Poria Cutis | FTIR-LC242 | 0.9016 | 0.7032 | 100% | 100% | |
| FTIR-LC242-210 | 0.905 | 0.698 | 100% | 100% | ||
| combination data of two medicinal parts | FTIR | 0.9548 | 0.8064 | 100% | 100% | |
| LC242 | 0.8147 | 0.6495 | 100% | 100% | ||
| LC210 | 0.6489 | 0.4806 | 94.23% | 88.46% | ||
| mid-level data fusion | Poria | FTIR-LC242 | 0.8266 | 0.5745 | 100% | 100% |
| FTIR-LC210 | 0.7453 | 0.5053 | 96.15% | 96.15% | ||
| FTIR-LC242-210 | 0.8286 | 0.5882 | 100% | 100% | ||
| Poria Cutis | FTIR-LC242 | 0.7386 | 0.5493 | 100% | 92.31% | |
| FTIR-LC210 | 0.7518 | 0.4991 | 100% | 96.15% | ||
| LC242-210 | 0.4617 | 0.228 | 76.92% | 73.08% | ||
| FTIR-LC242-210 | 0.7607 | 0.5558 | 100% | 96.15% | ||
| combination data of two medicinal parts | FTIR | 0.7564 | 0.5982 | 98.08% | 88.46% | |
| LC242 | 0.7761 | 0.4973 | 98.08% | 100% | ||
| LC210 | 0.676 | 0.3756 | 96.15% | 88.46% | ||
| Class | Location | Abbreviation | Elevation (m) | Latitude (°N) | Longitude (°E) | Parts | Sample Size |
|---|---|---|---|---|---|---|---|
| 1 | Beicheng Town, Hongta, Yuxi | BC | 1720 | 24.4319 | 102.5182 | inner part | 10 |
| epidermis | 10 | ||||||
| 2 | Tuodian Town, Shuangbai, Chuxiong | TD | 2062 | 24.6912 | 101.6493 | inner part | 10 |
| epidermis | 10 | ||||||
| 3 | Zhanhe Town, Ninglang, Lijiang | ZH | 2560 | 26.8832 | 100.9275 | inner part | 10 |
| epidermis | 10 | ||||||
| 4 | Dawen Town, Shuangjiang, Lincang | DW | 1438 | 23.3487 | 100.0047 | inner part | 10 |
| epidermis | 10 | ||||||
| 5 | Caodian Town, Yunlong, Dali | CD | 2066 | 25.6360 | 99.1320 | inner part | 10 |
| epidermis | 10 | ||||||
| 6 | Yongping Town, Jinggu, Pu’er | YP | 1077 | 23.4204 | 100.4044 | inner part | 10 |
| epidermis | 10 | ||||||
| 7 | Mengmeng Town, Shuangjiang, Lincang | MM | 1052 | 23.4779 | 99.8378 | inner part | 10 |
| epidermis | 10 | ||||||
| 8 | Baliu Town, Mojiang, Pu’er | BL | 1979 | 23.0676 | 101.9765 | inner part | 8 |
| epidermis | 8 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Q.-Q.; Huang, H.-Y.; Wang, Y.-Z. Geographical Authentication of Macrohyporia cocos by a Data Fusion Method Combining Ultra-Fast Liquid Chromatography and Fourier Transform Infrared Spectroscopy. Molecules 2019, 24, 1320. https://doi.org/10.3390/molecules24071320
Wang Q-Q, Huang H-Y, Wang Y-Z. Geographical Authentication of Macrohyporia cocos by a Data Fusion Method Combining Ultra-Fast Liquid Chromatography and Fourier Transform Infrared Spectroscopy. Molecules. 2019; 24(7):1320. https://doi.org/10.3390/molecules24071320
Chicago/Turabian StyleWang, Qin-Qin, Heng-Yu Huang, and Yuan-Zhong Wang. 2019. "Geographical Authentication of Macrohyporia cocos by a Data Fusion Method Combining Ultra-Fast Liquid Chromatography and Fourier Transform Infrared Spectroscopy" Molecules 24, no. 7: 1320. https://doi.org/10.3390/molecules24071320
APA StyleWang, Q. -Q., Huang, H. -Y., & Wang, Y. -Z. (2019). Geographical Authentication of Macrohyporia cocos by a Data Fusion Method Combining Ultra-Fast Liquid Chromatography and Fourier Transform Infrared Spectroscopy. Molecules, 24(7), 1320. https://doi.org/10.3390/molecules24071320