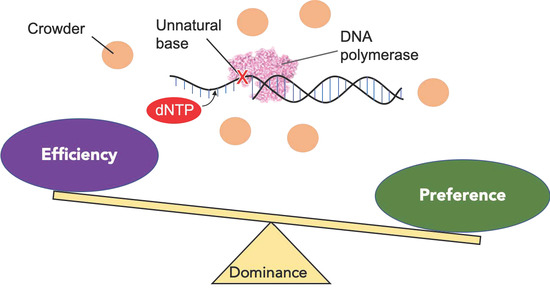
Effect of Molecular Crowding on DNA Polymerase Reactions along Unnatural DNA Templates
Abstract
:1. Introduction
2. Results and Discussion
2.1. Design of the Experimental Setup
2.2. Primer Extension in the Absence of the Crowder Molecule
2.3. Primer Extension under Crowding Conditions
3. Materials and Methods
3.1. Materials
3.2. Oligonucleotides
3.3. Primer Extension Assay
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Switzer, C.; Moroney, S.E.; Benner, S.A. Enzymatic incorporation of a new base pair into DNA and RNA. J. Am. Chem. Soc. 1989, 111, 8322–8323. [Google Scholar] [CrossRef]
- Kool, E.T. Hydrogen bonding, base stacking, and steric effects in DNA replication. Annu. Rev. Biophys. Biomol. Struct. 2001, 30, 1–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Turner, D.H.; Sugimoto, N.; Kierzek, R.; Dreiker, S.D. Free energy increments for hydrogen bonds in nucleic acid base pairs. J. Am. Chem. Soc. 1987, 109, 3783–3785. [Google Scholar] [CrossRef]
- Kool, E.T.; Morales, J.C.; Guckian, K.M. Mimicking the Structure and Function of DNA: Insights into DNA Stability and Replication. Angew. Chem. Int. Ed. 2000, 39, 990–1009. [Google Scholar] [CrossRef]
- Benner, S.A. Synthetic biology: Act natural. Nature 2003, 421, 118. [Google Scholar] [CrossRef] [PubMed]
- Hirao, I. Unnatural base pair systems for DNA/RNA-based biotechnology. Curr. Opin. Chem. Biol. 2006, 10, 622–627. [Google Scholar] [CrossRef] [PubMed]
- Ebel, S.; Lane, A.N.; Brown, T. Very stable mismatch duplexes: Structural and thermodynamic studies on tandem G.cntdot.A mismatches in DNA. Biochemistry 1992, 31, 12083–12086. [Google Scholar] [CrossRef]
- Carell, T.; Brandmayr, C.; Hienzsch, A.; Müller, M.; Pearson, D.; Reiter, V.; Thoma, I.; Thumbs, P.; Wagner, M. Structure and function of noncanonical nucleobases. Angew. Chem. Int. Ed. 2012, 51, 7110–7131. [Google Scholar] [CrossRef]
- Lu, H.; Krueger, A.T.; Gao, J.; Liu, H.; Kool, E.T. Toward a designed genetic system with biochemical function: Polymerase synthesis of single and multiple size-expanded DNA base pairs. Org. Biomol. Chem. 2010, 8, 2704–2710. [Google Scholar] [CrossRef] [Green Version]
- Malyshev, D.A.; Dhami, K.; Lavergne, T.; Chen, T.; Dai, N.; Foster, J.M.; Correa, I.R., Jr.; Romesberg, F.E. A semi-synthetic organism with an expanded genetic alphabet. Nature 2014, 509, 385–388. [Google Scholar] [CrossRef] [Green Version]
- Pezo, V.; Liu, F.W.; Abramov, M.; Froeyen, M.; Herdewijn, P.; Marlière, P. Binary Genetic Cassettes for Selecting XNA-Templated DNA Synthesis In Vivo. Angew. Chem. Int. Ed. 2013, 52, 8139–8143. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bande, O.; Abu El Asrar, R.; Braddick, D.; Dumbre, S.; Pezo, V.; Schepers, G.; Pinheiro, V.B.; Lescrinier, E.; Holliger, P.; Marlière, P.; et al. Isoguanine and 5-methyl-isocytosine bases, in vitro and in vivo. Chemistry 2015, 21, 5009–5022. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bande, O.; Braddick, D.; Agnello, S.; Jang, M.; Pezo, V.; Schepers, G.; Rozenski, J.; Lescrinier, E.; Marlière, P.; Herdewijn, P. Base pairing involving artificial bases in vitro and in vivo. Chem. Sci. 2016, 7, 995–1010. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nakano, S.; Miyoshi, D.; Sugimoto, N. Effects of molecular crowding on the structures, interactions, and functions of nucleic acids. Chem. Rev. 2014, 114, 2733–2758. [Google Scholar] [CrossRef]
- Nakano, S.-I.; Karimata, H.; Ohmichi, T.; Kawakami, J.; Sugimoto, N. The effect of molecular crowding with nucleotide length and cosolute structure on DNA duplex stability. J. Am. Chem. Soc. 2004, 126, 14330–14331. [Google Scholar] [CrossRef]
- Spink, C.H.; Chaires, J.B. Effects of hydration, ion release, and excluded volume on the melting of triplex and duplex DNA. Biochemistry 1999, 38, 496–508. [Google Scholar] [CrossRef]
- Ghosh, S.; Takahashi, S.; Ohyama, T.; Endoh, T.; Tateishi-Karimata, H.; Sugimoto, N. Nearest-neighbor parameters for predicting DNA duplex stability in diverse molecular crowding conditions. Proc. Natl. Acad. Sci. USA 2020, 117, 14194–14201. [Google Scholar] [CrossRef]
- Nakano, S.; Karimata, H.T.; Kitagawa, Y.; Sugimoto, N. Facilitation of RNA enzyme activity in the molecular crowding media of cosolutes. J. Am. Chem. Soc. 2009, 131, 16881–16888. [Google Scholar] [CrossRef]
- Nakano, S.I.; Sugimoto, N. The structural stability and catalytic activity of DNA and RNA oligonucleotides in the presence of organic solvents. Biophys. Rev. 2016, 8, 11–23. [Google Scholar] [CrossRef]
- Takahashi, S.; Okura, H.; Sugimoto, N. Bisubstrate Function of RNA Polymerases Triggered by Molecular Crowding Conditions. Biochemistry 2019, 58, 1081–1093. [Google Scholar] [CrossRef]
- Sismour, A.M.; Benner, S.A. The use of thymidine analogs to improve the replication of an extra DNA base pair: A synthetic biological system. Nucleic Acids Res. 2005, 33, 5640–5646. [Google Scholar] [CrossRef]
- Sasaki, Y.; Miyoshi, D.; Sugimoto, N. Effect of molecular crowding on DNA polymerase activity. Biotechnol. J. 2006, 1, 440–446. [Google Scholar] [CrossRef] [PubMed]
- Takahashi, S.; Okura, H.; Chilka, P.; Ghosh, S.; Sugimoto, N. Molecular crowding induces primer extension by RNA polymerase through base stacking beyond Watson–Crick rules. RSC Adv. 2020, 10, 33052–33058. [Google Scholar] [CrossRef]
- Takahashi, S.; Kim, K.T.; Podbevsek, P.; Plavec, J.; Kim, B.H.; Sugimoto, N. Recovery of the Formation and Function of Oxidized G-Quadruplexes by a Pyrene-Modified Guanine Tract. J. Am. Chem. Soc. 2018, 140, 5774–5783. [Google Scholar] [CrossRef] [PubMed]
Sample Availability: The plasmid DNA for the expression of KF is available from the authors. |





| Ino-DNA | Ino-HNA | Ino-AraNA | isoCMe-DNA | isoCMe-HNA | isoCMe-AraNA | isoG-DNA | isoG-HNA | |
|---|---|---|---|---|---|---|---|---|
| In the absence of PEG200 | ||||||||
| Efficiency (%) | 82.1 ± 3.3 | 83.9 ± 3.3 | 59.1 ± 0.6 | 31.5 ± 4.5 | 55.3 ± 6.0 | 19.0 ± 2.8 | 48.0 ± 3.6 | 60.0 ± 11.9 |
| (Preferred dNTP) | dCTP | dCTP | dCTP | dATP | dGTP | dATP | dTTP | dCTP |
| Preference (%) | 52.4 ± 0.5 | 40.0 ± 0.6 | 50.7 ± 0.9 | 75.9 ± 2.3 | 49.6 ± 5.7 | 68.4 ± 3.5 | 64.5 ± 4.5 | 48.7 ± 1.9 |
| In the presence of PEG200 | ||||||||
| Efficiency (%) | 62.1 ± 4.3 | 55.2 ± 3.5 | 64.5 ± 0.4 | 57.6 ± 3.8 | 64.0 ± 2.0 | 45.6 ± 11.1 | 57.2 ± 3.9 | 67.2 ± 6.4 |
| (Preferred dNTP) | dCTP | dCTP | dCTP | dATP | dGTP | dATP | dTTP | dCTP |
| Preference (%) | 32.6 ± 0.5 | 26.3 ± 0.6 | 42.9 ± 0.7 | 54.7 ± 2.3 | 36.9 ± 2.8 | 51.5 ± 3.6 | 43.3 ± 2.3 | 35.2 ± 0.6 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Takahashi, S.; Herdwijn, P.; Sugimoto, N. Effect of Molecular Crowding on DNA Polymerase Reactions along Unnatural DNA Templates. Molecules 2020, 25, 4120. https://doi.org/10.3390/molecules25184120
Takahashi S, Herdwijn P, Sugimoto N. Effect of Molecular Crowding on DNA Polymerase Reactions along Unnatural DNA Templates. Molecules. 2020; 25(18):4120. https://doi.org/10.3390/molecules25184120
Chicago/Turabian StyleTakahashi, Shuntaro, Piet Herdwijn, and Naoki Sugimoto. 2020. "Effect of Molecular Crowding on DNA Polymerase Reactions along Unnatural DNA Templates" Molecules 25, no. 18: 4120. https://doi.org/10.3390/molecules25184120
APA StyleTakahashi, S., Herdwijn, P., & Sugimoto, N. (2020). Effect of Molecular Crowding on DNA Polymerase Reactions along Unnatural DNA Templates. Molecules, 25(18), 4120. https://doi.org/10.3390/molecules25184120