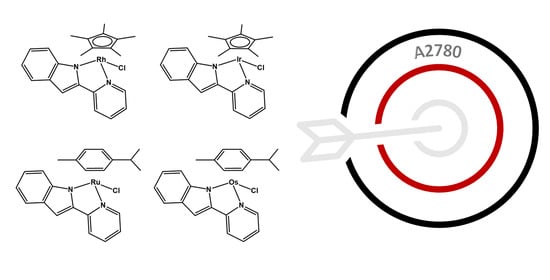
Synthesis, Characterisation and In Vitro Anticancer Activity of Catalytically Active Indole-Based Half-Sandwich Complexes
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis, Stability in Solution, Aquation and pKa Determination
2.2. Catalytic Reactions with Nicotinamide Adenine Dinucleotide
2.3. Chemosensitivity Assay
2.4. Reactions with Glutathione
2.5. Reactions with Nucleobases
3. Conclusions
4. Materials and Methods
4.1. Materials and Instrumentations
4.2. Synthesis
4.3. Solution Chemistry
4.4. Reaction with Nucleobases
4.5. Reaction with Glutathione
4.6. Oxidation of 1,4-NADH
4.7. NAD+ Reduction
4.8. Inhibition of NAD+ Reduction by Glutathione
4.9. Chemosensitivity Assays
4.10. Cell Viability Experiments
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- WHO. Fact Sheets. 2019. Available online: https://www.who.int/news (accessed on 1 October 2020).
- Aikman, B.; de Almeida, A.; Meier-Menches, S.M.; Casini, A. Aquaporins in cancer development: Opportunities for bioinorganic chemistry to contribute novel chemical probes and therapeutic agents. Metallomics 2018, 10, 696. [Google Scholar] [CrossRef] [Green Version]
- Shi, H.; Sadler, P.J. How promising is phototherapy for cancer? Br. J. Cancer 2020. [Google Scholar] [CrossRef] [PubMed]
- Laws, K.; Suntharalingam, K. The Next Generation of Anticancer Metallopharmaceuticals: Cancer Stem Cell-Active Inorganics. ChemBioChem 2018, 19, 2246. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.-Y.; Yi, Q.-Y.; Wang, Y.-J.; Du, F.; He, M.; Tang, B.; Wan, D.; Liu, Y.-J.; Huang, H.-L. Photoinduced anticancer activity studies of iridium(III) complexes targeting mitochondria and tubules. Eur. J. Med. Chem. 2018, 151, 568. [Google Scholar] [CrossRef] [PubMed]
- Pierroz, V.; Joshi, T.; Leonidova, A.; Mari, C.; Schur, J.; Ott, I.; Spiccia, L.; Ferrari, S.; Gasser, G. Molecular and Cellular Characterization of the Biological Effects of Ruthenium(II) Complexes Incorporating 2-Pyridyl-2-pyrimidine-4-carboxylic Acid. J. Am. Chem. Soc. 2012, 134, 20376. [Google Scholar] [CrossRef]
- Mulcahy, S.P.; Gründler, K.; Frias, C.; Wagner, L.; Prokop, A.; Meggers, E. Discovery of a strongly apoptotic ruthenium complex through combinatorial coordination chemistry. Dalton Trans. 2010, 39, 8177. [Google Scholar] [CrossRef] [Green Version]
- Velders, A.H.; Kooijman, H.; Spek, A.L.; Haasnoot, J.G.; de Vos, D.; Reedijk, J. Strong Differences in the in vitro Cytotoxicity of Three Isomeric Dichlorobis(2-phenylazopyridine)ruthenium(II) Complexes. Inorg. Chem. 2000, 39, 2966. [Google Scholar] [CrossRef]
- Hotze, A.C.G.; Velders, A.H.; Ugozzoli, F.; Biagini-Cingi, M.; Manotti-Lanfredi, A.M.; Haasnoot, J.G.; Reedijk, J. Synthesis, Characterization, and Crystal Structure of α-[Ru(azpy)2(NO3)2] (azpy = 2-(Phenylazo)pyridine) and the Products of Its Reactions with Guanine Derivatives. Inorg. Chem. 2000, 39, 3838. [Google Scholar] [CrossRef]
- Roy, S.; Colombo, E.; Vinck, R.; Mari, C.; Rubbiani, R.; Patra, M.; Gasser, G. Increased Lipophilicity of Halogenated Ruthenium(II) Polypyridyl Complexes Leads to Decreased Phototoxicity in vitro when Used as Photosensitizers for Photodynamic Therapy. ChemBioChem 2020. [Google Scholar] [CrossRef]
- Karges, J.; Yempala, T.; Tharaud, M.; Gibson, D.; Gasser, G. A Multi-action and Multi-target RuII–PtIV Conjugate Combining Cancer-Activated Chemotherapy and Photodynamic Therapy to Overcome Drug Resistant Cancers. Angew. Chem. Int. Ed. 2020, 59, 7069. [Google Scholar] [CrossRef]
- Zhang, W.-Y.; Banerjee, S.; Hughes, G.M.; Bridgewater, H.E.; Song, J.-I.; Breeze, B.G.; Clarkson, G.J.; Coverdale, J.P.C.; Sanchez-Cano, C.; Ponte, F.; et al. Ligand-centred redox activation of inert organoiridium anticancer catalysts. Chem. Sci. 2020, 11, 5466. [Google Scholar] [CrossRef]
- Renfrew, A.K.; Karges, J.; Scopelliti, R.; Bobbink, F.D.; Nowak-Sliwinska, P.; Gasser, G.; Dyson, P.J. Towards Light-Activated Ruthenium–Arene (RAPTA-Type) Prodrug Candidates. ChemBioChem 2019, 20, 2876. [Google Scholar] [CrossRef] [PubMed]
- Basu, U.; Karges, J.; Chotard, F.; Balan, C.; Le Gendre, P.; Gasser, G.; Bodio, E.; Malacea Kabbara, R. Investigation of photo-activation on ruthenium(II)–arene complexes for the discovery of potential selective cytotoxic agents. Polyhedron 2019, 172, 22. [Google Scholar] [CrossRef]
- Lord, R.M.; McGowan, P.C. Organometallic Iridium Arene Compounds: The Effects of C-Donor Ligands on Anticancer Activity. Chem. Lett. 2019, 48, 916. [Google Scholar] [CrossRef] [Green Version]
- Ang, W.H.; Casini, A.; Sava, G.; Dyson, P.J. Organometallic ruthenium-based antitumor compounds with novel modes of action. J. Organomet. Chem. 2011, 696, 989. [Google Scholar] [CrossRef]
- Casini, A.; Gabbiani, C.; Michelucci, E.; Pieraccini, G.; Moneti, G.; Dyson, P.J.; Messori, L. Exploring metallodrug–protein interactions by mass spectrometry: Comparisons between platinum coordination complexes and an organometallic ruthenium compound. J. Biol. Inorg. Chem. 2009, 14, 761. [Google Scholar] [CrossRef] [Green Version]
- Messori, L.; Orioli, P.; Vullo, D.; Alessio, E.; Iengo, E. A spectroscopic study of the reaction of NAMI, a novel ruthenium(III)anti-neoplastic complex, with bovine serum albumin. Eur. J. Biochem. 2000, 267, 1206. [Google Scholar] [CrossRef]
- Štarha, P.; Trávníček, Z. Non-platinum complexes containing releasable biologically active ligands. Coord. Chem. Rev. 2019, 395, 130. [Google Scholar] [CrossRef]
- Hudej, R.; Kljun, J.; Kandioller, W.; Repnik, U.; Turk, B.; Hartinger, C.G.; Keppler, B.K.; Miklavčič, D.; Turel, I. Synthesis and Biological Evaluation of the Thionated Antibacterial Agent Nalidixic Acid and Its Organoruthenium(II) Complex. Organometallics 2012, 31, 5867. [Google Scholar] [CrossRef]
- Aman, F.; Hanif, M.; Kubanik, M.; Ashraf, A.; Söhnel, T.; Jamieson, S.M.F.; Siddiqui, W.A.; Hartinger, C.G. Anti-Inflammatory Oxicams as Multi-donor Ligand Systems: pH- and Solvent-Dependent Coordination Modes of Meloxicam and Piroxicam to Ru and Os. Chem. Eur. J. 2017, 23, 4893. [Google Scholar] [CrossRef]
- Kljun, J.; León, I.E.; Peršič, Š.; Cadavid-Vargas, J.F.; Etcheverry, S.B.; He, W.; Bai, Y.; Turel, I. Synthesis and biological characterization of organoruthenium complexes with 8-hydroxyquinolines. J. Inorg. Biochem. 2018, 186, 187. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Patra, M.; Gasser, G. The medicinal chemistry of ferrocene and its derivatives. Nat. Rev. Chem. 2017, 1, 1–12. [Google Scholar] [CrossRef]
- Biancalana, L.; Batchelor, L.K.; De Palo, A.; Zacchini, S.; Pampaloni, G.; Dyson, P.J.; Marchetti, F. A general strategy to add diversity to ruthenium arene complexes with bioactive organic compounds via a coordinated (4-hydroxyphenyl)diphenylphosphine ligand. Dalton Trans. 2017, 46, 12001. [Google Scholar] [CrossRef] [PubMed]
- Banerjee, S.; Chakravarty, A.R. Metal Complexes of Curcumin for Cellular Imaging, Targeting, and Photoinduced Anticancer Activity. Acc. Chem. Res. 2015, 48, 2075. [Google Scholar] [CrossRef]
- Biot, C.; Nosten, F.; Fraisse, L.; Ter-Minassian, D.; Khalife, J.; Dive, D. The antimalarial ferroquine: From bench to clinic. Parasite 2011, 18, 207. [Google Scholar] [CrossRef] [Green Version]
- Wan, Y.; Li, Y.; Yan, C.; Yan, M.; Tang, Z. Indole: A privileged scaffold for the design of anti-cancer agents. Eur. J. Med. Chem. 2019, 183, 111691. [Google Scholar] [CrossRef]
- Dadashpour, S.; Emami, S. Indole in the target-based design of anticancer agents: A versatile scaffold with diverse mechanisms. Eur. J. Med. Chem. 2018, 150, 9. [Google Scholar] [CrossRef]
- Lal, S.; Snape, T.J. 2-Arylindoles: A Privileged Molecular Scaffold with Potent, Broad-Ranging Pharmacological Activity. Curr. Med. Chem. 2012, 19, 4828. [Google Scholar] [CrossRef]
- Hui, X.; Min, L. Developments of Indoles as Anti-HIV-1 Inhibitors. Curr. Pharm. Des. 2009, 15, 2120. [Google Scholar]
- Sang, Y.L.; Zhang, W.M.; Lv, P.C.; Zhu, H.L. Indole-based, antiproliferative agents targeting tubulin polymerization. Curr. Top. Med. Chem. 2017, 17, 120. [Google Scholar] [CrossRef]
- Grant, S.; Easley, C.; Kirkpatrick, P. Vorinostat. Nat. Rev. Drug Disc. 2007, 6, 21. [Google Scholar] [CrossRef] [PubMed]
- Furumai, R.; Matsuyama, A.; Kobashi, N.; Lee, K.H.; Nishiyama, M.; Nakajima, H.; Tanaka, A.; Komatsu, Y.; Nishino, N.; Yoshida, M.; et al. FK228 (depsipeptide) as a natural prodrug that inhibits class I histone deacetylases. Cancer Res. 2002, 62, 4916. [Google Scholar] [PubMed]
- Napper, A.D.; Hixon, J.; McDonagh, T.; Keavey, K.; Pons, J.-F.; Barker, J.; Yau, W.T.; Amouzegh, P.; Flegg, A.; Hamelin, E.; et al. Discovery of Indoles as Potent and Selective Inhibitors of the Deacetylase SIRT1. J. Med. Chem. 2005, 48, 8045. [Google Scholar] [CrossRef] [PubMed]
- Jordan, M.A.; Himes, R.H.; Wilson, L. Comparison of the Effects of Vinblastine, Vincristine, Vindesine, and Vinepidine on Microtubule Dynamics and Cell Proliferation. Cancer Res. 1985, 45, 2741. [Google Scholar]
- Almagro, L.; Fernández-Pérez, F.; Pedreño, M.A. Indole alkaloids from Catharanthus roseus: Bioproduction and their effect on human health. Molecules 2015, 20, 2973–3000. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bradner, W.T. Mitomycin C: A clinical update. Cancer Treat. Rev. 2001, 27, 35. [Google Scholar] [CrossRef] [Green Version]
- Soldevila-Barreda, J.J.; Sadler, P.J. Approaches to the design of catalytic metallodrugs. Curr. Opinion Chem. Biol. 2015, 25 (Suppl. C), 172. [Google Scholar] [CrossRef] [Green Version]
- Soldevila-Barreda, J.J.; Metzler-Nolte, N. Intracellular Catalysis with Selected Metal Complexes and Metallic Nanoparticles: Advances toward the Development of Catalytic Metallodrugs. Chem. Rev. 2019, 119, 829. [Google Scholar] [CrossRef]
- Alonso-de Castro, S.; Terenzi, A.; Gurruchaga-Pereda, J.; Salassa, L. Catalysis Concepts in Medicinal Inorganic Chemistry. Chem. Eur. J. 2019, 25, 6651. [Google Scholar] [CrossRef]
- Bose, S.; Ngo, A.H.; Do, L.H. Intracellular Transfer Hydrogenation Mediated by Unprotected Organoiridium Catalysts. J. Amer. Chem. Soc. 2017, 139, 8792. [Google Scholar] [CrossRef]
- Sasmal, P.K.; Carregal-Romero, S.; Han, A.A.; Streu, C.N.; Lin, Z.; Namikawa, K.; Elliott, S.L.; Köster, R.W.; Parak, W.J.; Meggers, E. Catalytic Azide Reduction in Biological Environments. ChemBioChem 2012, 13, 1116. [Google Scholar] [CrossRef] [PubMed]
- Martínez-Calvo, M.; Couceiro, J.R.; Destito, P.; Rodríguez, J.; Mosquera, J.; Mascareñas, J.L. Intracellular deprotection reactions mediated by palladium complexes equipped with designed phosphine ligands. ACS Catal. 2018, 8, 6055. [Google Scholar] [CrossRef] [PubMed]
- Völker, T.; Meggers, E. Transition-metal-mediated uncaging in living human cells—An emerging alternative to photolabile protecting groups. Curr. Opin. Chem. Biol. 2015, 25 (Suppl. C), 48. [Google Scholar] [CrossRef] [PubMed]
- Streu, C.; Meggers, E. Ruthenium-Induced Allylcarbamate Cleavage in Living Cells. Angew. Chem. Int. Ed. 2006, 45, 5645. [Google Scholar] [CrossRef]
- Völker, T.; Meggers, E. Chemical Activation in Blood Serum and Human Cell Culture: Improved Ruthenium Complex for Catalytic Uncaging of Alloc-Protected Amines. ChemBioChem 2017, 18, 1083. [Google Scholar] [CrossRef]
- Coverdale, J.P.C.; Romero-Canelón, I.; Sanchez-Cano, C.; Clarkson, G.J.; Habtemariam, A.; Wills, M.; Sadler, P.J. Asymmetric transfer hydrogenation by synthetic catalysts in cancer cells. Nat. Chem. 2018, 10, 347. [Google Scholar] [CrossRef]
- Chen, F.; Soldevila-Barreda, J.J.; Romero-Canelón, I.; Coverdale, J.P.C.; Song, J.I.; Clarkson, G.J.; Kasparkova, J.; Habtemariam, A.; Brabec, V.; Wolny, J.A.; et al. Effect of sulfonamidoethylenediamine substituents in RuII arene anticancer catalysts on transfer hydrogenation of coenzyme NAD+ by formate. Dalton Trans. 2018, 47, 7178. [Google Scholar] [CrossRef] [Green Version]
- Chen, F.; Romero-Canelón, I.; Soldevila-Barreda, J.J.; Song, J.I.; Coverdale, J.P.C.; Clarkson, G.J.; Kasparkova, J.; Habtemariam, A.; Wills, M.; Brabec, V.; et al. Transfer Hydrogenation and Antiproliferative Activity of Tethered Half-Sandwich Organoruthenium Catalysts. Organometallics 2018, 37, 1555. [Google Scholar] [CrossRef]
- Soldevila-Barreda, J.J.; Romero-Canelón, I.; Habtemariam, A.; Sadler, P.J. Transfer hydrogenation catalysis in cells as a new approach to anticancer drug design. Nat. Commun. 2015, 6, 6582. [Google Scholar] [CrossRef] [Green Version]
- Soldevila-Barreda, J.J.; Habtemariam, A.; Romero-Canelón, I.; Sadler, P.J. Half-sandwich rhodium(III) transfer hydrogenation catalysts: Reduction of NAD+ and pyruvate, and antiproliferative activity. J. Inorg. Biochem. 2015, 153, 322. [Google Scholar] [CrossRef] [Green Version]
- Liu, Z.; Romero-Canelón, I.; Qamar, B.; Hearn, J.M.; Habtemariam, A.; Barry, N.P.E.; Pizarro, A.M.; Clarkson, G.J.; Sadler, P.J. The Potent Oxidant Anticancer Activity of Organoiridium Catalysts. Angew. Chem. Int. Ed. 2014, 53, 3941. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Soldevila-Barreda, J.J.; Bruijnincx, P.C.A.; Habtemariam, A.; Clarkson, G.J.; Deeth, R.J.; Sadler, P.J. Improved Catalytic Activity of Ruthenium–Arene Complexes in the Reduction of NAD+. Organometallics 2012, 31, 5958. [Google Scholar] [CrossRef] [Green Version]
- Millett, A.J.; Habtemariam, A.; Romero-Canelón, I.; Clarkson, G.J.; Sadler, P.J. Contrasting Anticancer Activity of Half-Sandwich Iridium(III) Complexes Bearing Functionally Diverse 2-Phenylpyridine Ligands. Organometallics 2015, 34, 2683. [Google Scholar] [CrossRef] [PubMed]
- Betanzos-Lara, S.; Liu, Z.; Habtemariam, A.; Pizarro, A.M.; Qamar, B.; Sadler, P.J. Organometallic Ruthenium and Iridium Transfer-Hydrogenation Catalysts Using Coenzyme NADH as a Cofactor. Angew. Chem. Int. Ed. 2012, 51, 3897. [Google Scholar] [CrossRef] [PubMed]
- Matthew, C.K.; Ahern, K.G.; Van Holde, K.E. Biochemistry, 3rd ed.; Pearson Educacion: Madrid, Spain, 2002. [Google Scholar]
- Ying, W. NAD+/NADH and NADP+/NADPH in cellular functions and cell death: Regulation and biological consequences. Antioxid. Redox Signal. 2008, 10, 179. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Belenky, P.; Bogan, K.L.; Brenner, C. NAD+ metabolism in health and disease. Trends Biochem. Sci. 2007, 32, 12. [Google Scholar] [CrossRef]
- Hileman, E.O.; Liu, J.; Albitar, M.; Keating, M.J.; Huang, P. Intrinsic oxidative stress in cancer cells: A biochemical basis for therapeutic selectivity. Cancer Chemother. Pharm. 2004, 53, 209. [Google Scholar] [CrossRef]
- Liu, Z.; Sadler, P.J. Organoiridium Complexes: Anticancer Agents and Catalysts. Acc. Chem. Res. 2014, 47, 1174. [Google Scholar] [CrossRef]
- Meier-Menches, S.M.; Gerner, C.; Berger, W.; Hartinger, C.G.; Keppler, B.K. Structure–activity relationships for ruthenium and osmium anticancer agents—Towards clinical development. Chem. Soc. Rev. 2018, 47, 909. [Google Scholar] [CrossRef]
- Rilak Simović, A.; Masnikosa, R.; Bratsos, I.; Alessio, E. Chemistry and reactivity of ruthenium(II) complexes: DNA/protein binding mode and anticancer activity are related to the complex structure. Coord. Chem. Rev. 2019, 398, 113011. [Google Scholar] [CrossRef]
- Peacock, A.F.A.; Habtemariam, A.; Fernández, R.; Walland, V.; Fabbiani, F.P.A.; Parsons, S.; Aird, R.E.; Jodrell, D.I.; Sadler, P.J. Tuning the Reactivity of Osmium(II) and Ruthenium(II) Arene Complexes under Physiological Conditions. J. Am. Chem. Soc. 2006, 128, 1739. [Google Scholar] [CrossRef] [PubMed]
- Cross, J.M.; Gallagher, N.; Gill, J.H.; Jain, M.; McNeillis, A.W.; Rockley, K.L.; Tscherny, F.H.; Wirszycz, N.J.; Yufit, D.S.; Walton, J.W. Pyridylphosphinate metal complexes: Synthesis, structural characterisation and biological activity. Dalton Trans. 2016, 45, 12807. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Noffke, A.L.; Habtemariam, A.; Pizarro, A.M.; Sadler, P.J. Designing organometallic compounds for catalysis and therapy. Chem. Commun. 2012, 48, 5219. [Google Scholar] [CrossRef] [PubMed]
- de Torres, M.; Dimroth, J.; Arends, I.W.C.E.; Keilitz, J.; Hollmann, F. Towards Recyclable NAD(P)H Regeneration Catalysts. Molecules 2012, 17, 9835–9841. [Google Scholar] [CrossRef] [PubMed]
- Völker, T.; Dempwolff, F.; Graumann, P.L.; Meggers, E. Progress towards Bioorthogonal Catalysis with Organometallic Compounds. Angew. Chem. Int. Ed. 2014, 53, 10536. [Google Scholar] [CrossRef]
- Ngo, A.H.; Ibañez, M.; Do, L.H. Catalytic Hydrogenation of Cytotoxic Aldehydes Using Nicotinamide Adenine Dinucleotide (NADH) in Cell Growth Media. ACS Catal. 2016, 6, 2637. [Google Scholar] [CrossRef]
- Yang, L.; Bose, S.; Ngo, A.H.; Do, L.H. Innocent But Deadly: Nontoxic Organoiridium Catalysts Promote Selective Cancer Cell Death. ChemMedChem 2017, 12, 292. [Google Scholar] [CrossRef]
- Fu, Y.; Romero, M.J.; Habtemariam, A.; Snowden, M.E.; Song, L.; Clarkson, G.J.; Qamar, B.; Pizarro, A.M.; Unwin, P.R.; Sadler, P.J. The contrasting chemical reactivity of potent isoelectronic iminopyridine and azopyridine osmium(ii) arene anticancer complexes. Chem. Sci. 2012, 3, 2485. [Google Scholar] [CrossRef]
- Zhang, W.-Y.; Bridgewater, H.E.; Banerjee, S.; Soldevila-Barreda, J.J.; Clarkson, G.J.; Shi, H.; Imberti, C.; Sadler, P.J. Ligand-Controlled Reactivity and Cytotoxicity of Cyclometalated Rhodium(III) Complexes. Eur. J. Inorg. Chem. 2019, 2020, 1052. [Google Scholar] [CrossRef]
- Lo, H.C.; Buriez, O.; Kerr, J.B.; Fish, R.H. Regioselective reduction of NAD+ models with [Cp*Rh(bpy)H]+: Structure-activity relationships and mechanistic aspects in the formation of the 1,4-NADH derivatives. Angew. Chem. Int. Ed. 1999, 38, 1429. [Google Scholar] [CrossRef]
- Lo, H.C.; Leiva, C.; Buriez, O.; Kerr, J.B.; Olmstead, M.M.; Fish, R.H. Bioorganometallic chemistry. 13. Regioselective reduction of NAD+ models, 1-benzylnicotinamde triflate and Β-nicotinamide ribose-5′-methyl phosphate, with in situ generated [Cp*Rh(Bpy)H]+: Structure-activity relationships, kinetics, and mechanistic aspects in the formation of the 1,4-NADH derivatives. Inorg. Chem. 2001, 40, 6705. [Google Scholar] [PubMed]
- Haquette, P.; Talbi, B.; Barilleau, L.; Madern, N.; Fosse, C.; Salmain, M. Chemically engineered papain as artificial formate dehydrogenase for NAD(P)H regeneration. Org. Biomol. Chem. 2011, 9, 5720. [Google Scholar] [CrossRef] [PubMed]
- Canivet, J.; Süss-Fink, G.; Štěpnička, P. Water-Soluble Phenanthroline Complexes of Rhodium, Iridium and Ruthenium for the Regeneration of NADH in the Enzymatic Reduction of Ketones. Eur. J. Inorg. Chem. 2007, 4736. [Google Scholar] [CrossRef]
- Soldevila-Barreda, J.J.; Azmanova, M.; Pitto-Barry, A.; Cooper, P.A.; Shnyder, S.D.; Barry, N.P.E. Preclinical Anticancer Activity of an Electron-Deficient Organoruthenium(II) Complex. ChemMedChem 2020, 15, 982. [Google Scholar] [CrossRef] [PubMed]
- Azmanova, M.; Soldevila-Barreda, J.; Bani Hani, H.; Lord, R.M.; Pitto-Barry, A.; Picksley, S.M.; Barry, N.P.E. Anticancer Activity of Electron-Deficient Metal Complexes against Colorectal Cancer in vitro Models. ChemMedChem 2019, 14, 1887. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, K.; Jakob, U. The role of thiols in antioxidant systems. Free Radic. Biol. Med. 2019, 140, 14. [Google Scholar] [CrossRef]
- Lushchak, V.I. Glutathione Homeostasis and Functions: Potential Targets for Medical Interventions. Amino Acids 2012, 2012, 1. [Google Scholar] [CrossRef] [Green Version]
- Rocha, C.R.R.; Garcia, C.C.M.; Vieira, D.B.; Quinet, A.; de Andrade-Lima, L.C.; Munford, V.; Belizario, J.E.; Menck, C.F.M. Glutathione depletion sensitizes cisplatin- and temozolomide-resistant glioma cells in vitro and in vivo. Cell Death Discov. 2014, 5, e1505. [Google Scholar] [CrossRef]
- Kartalou, M.; Essigmann, J.M. Mechanisms of resistance to cisplatin. Muta. Res. 2001, 478, 23. [Google Scholar] [CrossRef]
- Wang, F.; Xu, J.; Wu, K.; Weidt, S.K.; Mackay, C.L.; Langridge-Smith, P.R.R.; Sadler, P.J. Competition between glutathione and DNA oligonucleotides for ruthenium(ii) arene anticancer complexes. Dalton Trans. 2013, 42, 3188. [Google Scholar] [CrossRef]
- Li, J.; Tian, Z.; Ge, X.; Xu, Z.; Feng, Y.; Liu, Z. Design, synthesis, and evaluation of fluorine and Naphthyridine–Based half-sandwich organoiridium/ruthenium complexes with bioimaging and anticancer activity. Eur. J. Med. Chem. 2019, 163, 830. [Google Scholar] [CrossRef] [PubMed]
- Purkait, K.; Ruturaj; Mukherjee, A.; Gupta, A. ATP7B Binds Ruthenium(II) p-Cymene Half-Sandwich Complexes: Role of Steric Hindrance and Ru–I Coordination in Rescuing the Sequestration. Inorg. Chem. 2019, 58, 15659. [Google Scholar] [CrossRef] [PubMed]
- Pages, B.J.; Ang, D.L.; Wright, E.P.; Aldrich-Wright, J.R. Metal complex interactions with DNA. Dalton Trans. 2015, 44, 3505. [Google Scholar] [CrossRef] [PubMed]
- Baik, M.-H.; Friesner, R.A.; Lippard, S.J. Theoretical Study of Cisplatin Binding to Purine Bases: Why Does Cisplatin Prefer Guanine over Adenine? J. Am. Chem. Soc. 2003, 125, 14082. [Google Scholar] [CrossRef] [PubMed]
- Thordarson, P. Determining association constants from titration experiments in supramolecular chemistry. Chem. Soc. Rev. 2011, 40, 1305. [Google Scholar] [CrossRef] [PubMed]
- Gutten, O.; Rulíšek, L. Predicting the Stability Constants of Metal-Ion Complexes from First Principles. Inor. Chem. 2013, 52, 10347. [Google Scholar] [CrossRef]
- Barry, N.P.E.; Furrer, J.; Therrien, B. In- and Out-of-Cavity Interactions by Modulating the Size of Ruthenium Metallarectangles. Helv. Chim. Acta 2010, 93, 1313. [Google Scholar] [CrossRef] [Green Version]
- Barry, N.P.E.; Furrer, J.; Freudenreich, J.; Süss-Fink, G.; Therrien, B. Designing the Host-Guest Properties of Tetranuclear Arene Ruthenium Metalla-Rectangles to Accommodate a Pyrene Molecule. Eur. J. Inorg. Chem. 2010, 2010, 725. [Google Scholar] [CrossRef] [Green Version]
- Barry, N.P.E.; Therrien, B. Host-Guest Chemistry in the Hexanuclear (Arene)ruthenium Metalla-Prismatic Cage Ru-6(p-cymene)(6)(tpt)(2)(dhnq)(3) (6+). Eur. J. Inorg. Chem. 2009, 31, 4695. [Google Scholar] [CrossRef] [Green Version]
Sample Availability: Samples of the compounds are available from the authors. |






| Compound | pKa a | K9-EtG (M−1) b | TOF (h−1) c |
|---|---|---|---|
| [Cp*Rh(ind-py)Cl] (1) | - | 25.6 × 103 | 14.6 ± 0.7 |
| [Cp*Ir(ind-py)Cl] (2) | 9.59 ± 0.09 (λ = 300 nm) | - | - |
| [(p-cym)Ru(ind-py)Cl] (3) | 10.63 ± 0.03 (λ = 255 nm) | 71.6 × 103 | 7.95 ± 0.81 |
| [(p-cym)Os(ind-py)Cl] (4) | 10.15 ± 0.04 (λ = 355 nm) | 95.4 × 103 | 5.74 ± 0.35 |
| IC50 Values (µM) | |||
|---|---|---|---|
| Compound | A2780 | A2780cisR | PNT2 |
| [Cp*Rh(ind-py)Cl] (1) | 13.0 ± 1.7 | 21.5 ± 3.5 | 34.7 ± 6.1 |
| [Cp*Ir(ind-py)Cl] (2) | 22.0 ± 2.5 | 32.7 ± 1.7 | 32.1 ± 3.9 |
| [(p-cym)Ru(ind-py)Cl] (3) | 47.3 ± 4.7 | 62.8 ± 5.3 | 54.3 ± 5.4 |
| [(p-cym Os)(ind-py)Cl] (4) | 18.8 ± 0.3 | 21.5 ± 1.3 | 34.2 ± 2.2 |
| Ind-py | >100 | >100 | >100 |
| Cisplatin | 10.3 ± 0.5 | 22.4 ± 0.5 | 43 ± 3 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Soldevila-Barreda, J.J.; Fawibe, K.B.; Azmanova, M.; Rafols, L.; Pitto-Barry, A.; Eke, U.B.; Barry, N.P.E. Synthesis, Characterisation and In Vitro Anticancer Activity of Catalytically Active Indole-Based Half-Sandwich Complexes. Molecules 2020, 25, 4540. https://doi.org/10.3390/molecules25194540
Soldevila-Barreda JJ, Fawibe KB, Azmanova M, Rafols L, Pitto-Barry A, Eke UB, Barry NPE. Synthesis, Characterisation and In Vitro Anticancer Activity of Catalytically Active Indole-Based Half-Sandwich Complexes. Molecules. 2020; 25(19):4540. https://doi.org/10.3390/molecules25194540
Chicago/Turabian StyleSoldevila-Barreda, Joan J., Kehinde B. Fawibe, Maria Azmanova, Laia Rafols, Anaïs Pitto-Barry, Uche B. Eke, and Nicolas P. E. Barry. 2020. "Synthesis, Characterisation and In Vitro Anticancer Activity of Catalytically Active Indole-Based Half-Sandwich Complexes" Molecules 25, no. 19: 4540. https://doi.org/10.3390/molecules25194540
APA StyleSoldevila-Barreda, J. J., Fawibe, K. B., Azmanova, M., Rafols, L., Pitto-Barry, A., Eke, U. B., & Barry, N. P. E. (2020). Synthesis, Characterisation and In Vitro Anticancer Activity of Catalytically Active Indole-Based Half-Sandwich Complexes. Molecules, 25(19), 4540. https://doi.org/10.3390/molecules25194540