Synthesis of Ferulenol by Engineered Escherichia coli: Structural Elucidation by Using the In Silico Tools
Abstract
:1. Introduction
2. Results
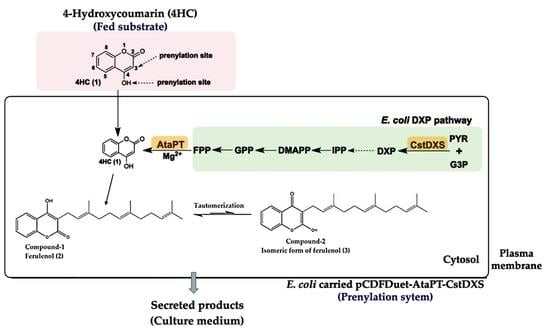
2.1. Construction of Plasmid pCDFDuet-AtaPT-CstDXS
2.2. Identification of Prenylated 4HC Derivative Using LC-ESI(-)-QTOF-MS/MS
2.3. Structural Annotation of Compound-1 by Using MetFrag Web Service
2.4. Computationally Assisted Identification of the Prenylated Product by Using SIRIUS (Version 4.8.2)
2.5. Structural Annotation and Compound Classification of Compound-1 by Using CSI:FIngerID and CANOPUS
2.6. Identification of Farnesylated-4HC Derivatives by Using LC-QTOF-MS/MS in Positive ESI Mode
3. Discussion
4. Materials and Methods
4.1. Reagents
4.2. Construction of Plasmid pCDFDuet-AtaPT-CstDXS
4.3. Bioconversion of 4HC (1) into the Farnesylated-4HC Derivatives
4.4. Extraction of Prenylated Products from the Culture Medium
4.5. Identification of Metabolites by LC-ESI-QTOF-MS/MS
4.6. Structural Annotation of Prenylated Products by Using MetFrag
4.7. Structural Annotations of Prenylated 4HC Analog Using SIRIUS Tool
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Yazaki, K.; Sasaki, K.; Tsurumaru, Y. Prenylation of aromatic compounds, a key diversification of plant secondary metabolites. Phytochemistry 2009, 70, 1739–1745. [Google Scholar] [CrossRef] [Green Version]
- Botta, B.; Vitali, A.; Menendez, P.; Misiti, D.; Monache, G. Prenylated Flavonoids: Pharmacology and Biotechnology. Curr. Med. Chem. 2005, 12, 713–739. [Google Scholar] [CrossRef] [PubMed]
- Alhassan, A.M.; Abdullahi, M.I.; Uba, A.; Umar, A. Prenylation of aromatic secondary metabolites: A new frontier for development of novel drugs. Trop. J. Pharm. Res. 2014, 13, 307–314. [Google Scholar] [CrossRef]
- Bartmańska, A.; Tronina, T.; Popłoński, J.; Milczarek, M.; Filip-Psurska, B.; Wietrzyk, J. Highly cancer selective antiproliferative activity of natural prenylated flavonoids. Molecules 2018, 23, 2922. [Google Scholar] [CrossRef] [Green Version]
- Akaberi, M.; Iranshahy, M.; Iranshahi, M. Review of the traditional uses, phytochemistry, pharmacology and toxicology of giant fennel (Ferula communis l. subsp. communis). Iran. J. Basic Med. Sci. 2015, 18, 1050–1062. [Google Scholar] [PubMed]
- Trantas, E.A.; Koffas, M.A.G.; Xu, P.; Ververidis, F. When plants produce not enough or at all: Metabolic engineering of flavonoids in microbial hosts. Front. Plant Sci. 2015, 6, 1–16. [Google Scholar] [CrossRef] [Green Version]
- Ajikumar, P.K.; Tyo, K.; Carlsen, S.; Mucha, O.; Phon, T.H.; Stephanopoulos, G. Terpenoids: Opportunities for biosynthesis of natural product drugs using engineered microorganisms. Mol. Pharm. 2008, 5, 167–190. [Google Scholar] [CrossRef]
- Shen, R.; Jiang, X.; Ye, W.; Song, X.; Liu, L.; Lao, X.; Wu, C. A novel and practical synthetic route for the total synthesis of lycopene. Tetrahedron 2011, 67, 5610–5614. [Google Scholar] [CrossRef]
- Zhou, K.; Edgar, S.; Stephanopoulos, G. Engineering Microbes to Synthesize Plant Isoprenoids. Methods Enzymol. 2016, 575, 225–245. [Google Scholar]
- Gliszczyńska, A.; Brodelius, P.E. Sesquiterpene coumarins. Phytochem. Rev. 2011, 11, 77–96. [Google Scholar] [CrossRef]
- Bocca, C.; Gabriel, L.; Bozzo, F.; Miglietta, A. Microtubule-interacting activity and cytotoxicity of the prenylated coumarin ferulenol. Planta Med. 2002, 68, 1135–1137. [Google Scholar] [CrossRef]
- Altmann, K.H.; Gertsch, J. Anticancer drugs from nature - Natural products as a unique source of new microtubule-stabilizing agents. Nat. Prod. Rep. 2007, 24, 327–357. [Google Scholar] [CrossRef] [PubMed]
- Lariche, N.; Lahouel, M.; Benguedouar, L.; Zellagui, A. Ferulenol, a sesquiterpene coumarin, induce apoptosis via mitochondrial dysregulation in lung cancer induced by benzo[a]pyrene: Involvement of Bcl2 protein. Anticancer. Agents Med. Chem. 2017, 17, 1357–1362. [Google Scholar] [CrossRef]
- Wiart, C. Lead Compounds from Medicinal Plants for the Treatment of Cancer, 1st ed.; Academic Press: Cambridge, MA, USA, 2012; ISBN 9780123983824.
- Gebauer, M. Synthesis and structure-activity relationships of novel warfarin derivatives. Bioorganic Med. Chem. 2007, 15, 2414–2420. [Google Scholar] [CrossRef]
- Monti, M.; Pinotti, M.; Appendino, G.; Dallocchio, F.; Bellini, T.; Antognoni, F.; Poli, F.; Bernardi, F. Characterization of anti-coagulant properties of prenylated coumarin ferulenol. Biochim. Biophys. Acta Gen. Subj. 2007, 1770, 1437–1440. [Google Scholar] [CrossRef] [PubMed]
- Appendino, G.; Mercalli, E.; Fuzzati, N.; Arnoldi, L.; Stavri, M.; Gibbons, S.; Ballero, M.; Maxia, A. Antimycobacterial coumarins from the Sardinian giant fennel (Ferula communis). J. Nat. Prod. 2004, 67, 2108–2110. [Google Scholar] [CrossRef] [PubMed]
- de Bruijn, W.J.C.; Levisson, M.; Beekwilder, J.; van Berkel, W.J.H.; Vincken, J.P. Plant Aromatic Prenyltransferases: Tools for Microbial Cell Factories. Trends Biotechnol. 2020, 38, 917–934. [Google Scholar] [CrossRef]
- Ward, V.C.A.; Chatzivasileiou, A.O.; Stephanopoulos, G. Metabolic engineering of Escherichia coli for the production of isoprenoids. FEMS Microbiol. Lett. 2018, 365, 1–9. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhao, Y.; Yang, J.; Qin, B.; Li, Y.; Sun, Y.; Su, S.; Xian, M. Biosynthesis of isoprene in Escherichia coli via methylerythritol phosphate (MEP) pathway. Appl. Microbiol. Biotechnol. 2011, 90, 1915–1922. [Google Scholar] [CrossRef]
- Volke, D.C.; Rohwer, J.; Fischer, R.; Jennewein, S. Investigation of the methylerythritol 4-phosphate pathway for microbial terpenoid production through metabolic control analysis. Microb. Cell Fact. 2019, 18, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Niu, F.X.; Lu, Q.; Bu, Y.F.; Liu, J.Z. Metabolic engineering for the microbial production of isoprenoids: Carotenoids and isoprenoid-based biofuels. Synth. Syst. Biotechnol. 2017, 2, 167–175. [Google Scholar] [CrossRef]
- Li, Y.; Wang, G. Strategies of isoprenoids production in engineered bacteria. J. Appl. Microbiol. 2016, 121, 932–940. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yuan, L.Z.; Rouvière, P.E.; LaRossa, R.A.; Suh, W. Chromosomal promoter replacement of the isoprenoid pathway for enhancing carotenoid production in E. coli. Metab. Eng. 2006, 8, 79–90. [Google Scholar] [CrossRef]
- Takahashi, H.; Aihara, Y.; Ogawa, Y.; Murata, Y.; Nakajima, K.-I.; Iida, M.; Shirai, M.; Fujisaki, S. Suppression of phenotype of Escherichia coli mutant defective in farnesyl diphosphate synthase by overexpression of gene for octaprenyl diphosphate synthase. Biosci. Biotechnol. Biochem. 2018, 82, 1003–1010. [Google Scholar] [CrossRef] [Green Version]
- Cao, Y.; Zhang, R.; Liu, W.; Zhao, G.; Niu, W.; Guo, J.; Xian, M.; Liu, H. Manipulation of the precursor supply for high-level production of longifolene by metabolically engineered Escherichia coli. Sci. Rep. 2019, 9, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Lee, P.C.; Petri, R.; Mijts, B.N.; Watts, K.T.; Schmidt-Dannert, C. Directed evolution of Escherichia coli farnesyl diphosphate synthase (IspA) reveals novel structural determinants of chain length specificity. Metab. Eng. 2005, 7, 18–26. [Google Scholar] [CrossRef]
- Vallon, T.; Ghanegaonkar, S.; Vielhauer, O.; Müller, A.; Albermann, C.; Sprenger, G.; Reuss, M.; Lemuth, K. Quantitative analysis of isoprenoid diphosphate intermediates in recombinant and wild-type Escherichia coli strains. Appl. Microbiol. Biotechnol. 2008, 81, 175–182. [Google Scholar] [CrossRef]
- Boghigian, B.A.; Salas, D.; Ajikumar, P.K.; Stephanopoulos, G.; Pfeifer, B.A. Analysis of heterologous taxadiene production in K- and B-derived Escherichia coli. Appl. Microbiol. Biotechnol. 2012, 93, 1651–1661. [Google Scholar] [CrossRef] [PubMed]
- Huang, Q.; Roessner, C.A.; Croteau, R.; Scott, A.I. Engineering Escherichia coli for the synthesis of taxadiene, a key intermediate in the biosynthesis of taxol. Bioorganic Med. Chem. 2001, 9, 2237–2242. [Google Scholar] [CrossRef]
- Albrecht, M.; Misawa, N.; Sandmann, G. Metabolic engineering of the terpenoid biosynthetic pathway of Escherichia coli for production of the carotenoids β-carotene and zeaxanthin. Biotechnol. Lett. 1999, 21, 791–795. [Google Scholar] [CrossRef]
- Schmidt-Dannert, C. Engineering novel carotenoids in microorganisms. Curr. Opin. Biotechnol. 2000, 11, 255–261. [Google Scholar] [CrossRef]
- Tsuruta, H.; Paddon, C.J.; Eng, D.; Lenihan, J.R.; Horning, T.; Anthony, L.C.; Regentin, R.; Keasling, J.D.; Renninger, N.S.; Newman, J.D. High-level production of amorpha-4, 11-diene, a precursor of the antimalarial agent artemisinin, in Escherichia coli. PLoS ONE 2009, 4, e4489. [Google Scholar] [CrossRef] [PubMed]
- Anthony, J.R.; Anthony, L.C.; Nowroozi, F.; Kwon, G.; Newman, J.D.; Keasling, J.D. Optimization of the mevalonate-based isoprenoid biosynthetic pathway in Escherichia coli for production of the anti-malarial drug precursor amorpha-4,11-diene. Metab. Eng. 2009, 11, 13–19. [Google Scholar] [CrossRef] [PubMed]
- Sasaki, K.; Tsurumaru, Y.; Yazakiy, K. Prenylation of flavonoids by biotransformation of yeast expressing plant membrane-bound prenyltransferase SfN8DT-1. Biosci. Biotechnol. Biochem. 2009, 73, 759–761. [Google Scholar] [CrossRef] [Green Version]
- Luo, X.; Reiter, M.A.; d’Espaux, L.; Wong, J.; Denby, C.M.; Lechner, A.; Zhang, Y.; Grzybowski, A.T.; Harth, S.; Lin, W.; et al. Complete biosynthesis of cannabinoids and their unnatural analogues in yeast. Nature 2019, 567, 123–126. [Google Scholar] [CrossRef]
- Li, Z.; Zhao, G.; Liu, H.; Guo, Y.; Wu, H.; Sun, X.; Wu, X.; Zheng, Z. Biotransformation of menadione to its prenylated derivative MK-3 using recombinant Pichia pastoris. J. Ind. Microbiol. Biotechnol. 2017, 44, 973–985. [Google Scholar] [CrossRef]
- Ma, Y.; McClure, D.D.; Somerville, M.V.; Proschogo, N.W.; Dehghani, F.; Kavanagh, J.M.; Coleman, N.V. Metabolic Engineering of the MEP Pathway in Bacillus subtilis for increased biosynthesis of menaquinone-7. ACS Synth. Biol. 2019, 8, 1620–1630. [Google Scholar] [CrossRef] [PubMed]
- Heide, L. Prenyl transfer to aromatic substrates: Genetics and enzymology. Curr. Opin. Chem. Biol. 2009, 13, 171–179. [Google Scholar] [CrossRef]
- Zhou, K.; Ludwig, L.; Li, S.M. Friedel-Crafts alkylation of acylphloroglucinols catalyzed by a fungal indole prenyltransferase. J. Nat. Prod. 2015, 78, 929–933. [Google Scholar] [CrossRef]
- Liebhold, M.; Xie, X.; Li, S.M. Expansion of enzymatic Friedel-Crafts alkylation on indoles: Acceptance of unnatural β-unsaturated allyl diphospates by dimethylallyl-tryptophan synthases. Org. Lett. 2012, 14, 4882–4885. [Google Scholar] [CrossRef]
- Fan, A.; Xie, X.; Li, S.M. Tryptophan prenyltransferases showing higher catalytic activities for Friedel-Crafts alkylation of o- and m-tyrosines than tyrosine prenyltransferases. Org. Biomol. Chem. 2015, 13, 7551–7557. [Google Scholar] [CrossRef] [Green Version]
- Miranda, C.L.; Aponso, G.L.M.; Stevens, J.F.; Deinzer, M.L.; Buhler, D.R. Prenylated chalcones and flavanones as inducers of quinone reductase in mouse Hepa 1c1c7 cells. Cancer Lett. 2000, 149, 21–29. [Google Scholar] [CrossRef]
- Dong, X.; Fan, Y.; Yu, L.; Hu, Y. Synthesis of four natural prenylflavonoids and their estrogen-like activities. Arch. Pharm. (Weinheim). 2007, 340, 372–376. [Google Scholar] [CrossRef] [PubMed]
- Appendino, G.; Gibbons, S.; Giana, A.; Pagani, A.; Grassi, G.; Stavri, M.; Smith, E.; Rahman, M.M. Antibacterial cannabinoids from Cannabis sativa: A structure-activity study. J. Nat. Prod. 2008, 71, 1427–1430. [Google Scholar] [CrossRef] [PubMed]
- Chen, R.; Gao, B.; Liu, X.; Ruan, F.; Zhang, Y.; Lou, J.; Feng, K.; Wunsch, C.; Li, S.M.; Dai, J.; et al. Molecular insights into the enzyme promiscuity of an aromatic prenyltransferase. Nat. Chem. Biol. 2017, 13, 226–234. [Google Scholar] [CrossRef] [PubMed]
- Abdou, M.M.; El-Saeed, R.A.; Bondock, S. Recent advances in 4-hydroxycoumarin chemistry. Part 1: Synthesis and reactions. Arab. J. Chem. 2019, 12, 88–121. [Google Scholar] [CrossRef] [Green Version]
- Sitthithaworn, W.; Wungsintaweekul, J.; Sirisuntipong, T.; Charoonratana, T.; Ebizuka, Y.; De-Eknamkul, W. Cloning and expression of 1-deoxy-d-xylulose 5-phosphate synthase cDNA from Croton stellatopilosus and expression of 2C-methyl-d-erythritol 4-phosphate synthase and geranylgeranyl diphosphate synthase, key enzymes of plaunotol biosynthesis. J. Plant Physiol. 2010, 167, 292–300. [Google Scholar] [CrossRef]
- Xiao, J.F.; Zhou, B.; Ressom, H.W. Metabolite identification and quantitation in LC-MS/MS-based metabolomics. TrAC - Trends Anal. Chem. 2012, 32, 1–14. [Google Scholar] [CrossRef] [Green Version]
- Zhou, B.; Xiao, J.F.; Tuli, L.; Ressom, H.W. LC-MS-based metabolomics. Mol. Biosyst. 2012, 8, 470–481. [Google Scholar] [CrossRef] [Green Version]
- Blaženović, I.; Kind, T.; Ji, J.; Fiehn, O. Software tools and approaches for compound identification of LC-MS/MS data in metabolomics. Metabolites 2018, 8, 31. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ruttkies, C.; Schymanski, E.L.; Wolf, S.; Hollender, J.; Neumann, S. MetFrag relaunched: Incorporating strategies beyond in silico fragmentation. J. Cheminform. 2016, 8, 1–16. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wolf, S.; Schmidt, S.; Müller-Hannemann, M.; Neumann, S. In silico fragmentation for computer assisted identification of metabolite mass spectra. BMC Bioinform. 2010, 11, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Dührkop, K.; Fleischauer, M.; Ludwig, M.; Aksenov, A.A.; Melnik, A.V.; Meusel, M.; Dorrestein, P.C.; Rousu, J.; Böcker, S. SIRIUS 4: A rapid tool for turning tandem mass spectra into metabolite structure information. Nat. Methods 2019, 16, 299–302. [Google Scholar] [CrossRef] [Green Version]
- Dührkop, K.; Shen, H.; Meusel, M.; Rousu, J.; Böcker, S. Searching molecular structure databases with tandem mass spectra using CSI:FingerID. Proc. Natl. Acad. Sci. USA 2015, 112, 12580–12585. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dührkop, K.; Nothias, L.F.; Fleischauer, M.; Ludwig, M.; Hoffmann, M.A.; Rousu, J.; Dorrestein, P.C.; Böcker, S. Classes for the masses: Systematic classification of unknowns using fragmentation spectra. bioRxiv 2020, 046672. [Google Scholar] [CrossRef] [Green Version]
- Fourel, I.; Hugnet, C.; Goy-Thollot, I.; Berny, P. Validation of a new liquid chromatography-tandem mass spectrometry ion-trap technique for the simultaneous determination of thirteen anticoagulant rodenticides, drugs, or natural products. J. Anal. Toxicol. 2010, 34, 95–102. [Google Scholar] [CrossRef] [Green Version]
- Klamrak, A.; Nabnueangsap, J.; Nualkaew, N. Biotransformation of benzoate to 2,4,6-trihydroxybenzophenone by engineered Escherichia coli. Molecules 2021, 26, 2779. [Google Scholar] [CrossRef]
- Cortés, I.; Cala, L.J.; Bracca, A.B.J.; Kaufman, T.S. Furo[3,2-c]coumarins carrying carbon substituents at C-2 and/or C-3. Isolation, biological activity, synthesis and reaction mechanisms. RSC Adv. 2020, 10, 33344–33377. [Google Scholar] [CrossRef]
- Lee, P.C.; Schmidt-Dannert, C. Metabolic engineering towards biotechnological production of carotenoids in microorganisms. Appl. Microbiol. Biotechnol. 2002, 60, 1–11. [Google Scholar]
- Lin, Y.; Shen, X.; Yuan, Q.; Yan, Y. Microbial biosynthesis of the anticoagulant precursor 4-hydroxycoumarin. Nat. Commun. 2013, 4, 1–8. [Google Scholar] [CrossRef] [Green Version]
- Gustafsson, C.; Govindarajan, S.; Minshull, J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004, 22, 346–353. [Google Scholar] [CrossRef]
- Jones, K.L.; Kim, S.W.; Keasling, J.D. Low-copy plasmids can perform as well as or better than high-copy plasmids for metabolic engineering of bacteria. Metab. Eng. 2000, 2, 328–338. [Google Scholar] [CrossRef] [PubMed]
- Singh, A.; Upadhyay, V.; Upadhyay, A.K.; Singh, S.M.; Panda, A.K. Protein recovery from inclusion bodies of Escherichia coli using mild solubilization process. Microb. Cell Fact. 2015, 14, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Tolia, N.H.; Joshua-Tor, L. Strategies for protein coexpression in Escherichia coli. Nat. Methods 2006, 3, 55–64. [Google Scholar] [CrossRef] [PubMed]
- Ridder, L.; Van Der Hooft, J.J.J.; Verhoeven, S.; De Vos, R.C.H.; Bino, R.J.; Vervoort, J. Automatic chemical structure annotation of an LC-MSn based metabolic profile from green tea. Anal. Chem. 2013, 85, 6033–6040. [Google Scholar] [CrossRef] [PubMed]







Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Klamrak, A.; Nabnueangsap, J.; Puthongking, P.; Nualkaew, N. Synthesis of Ferulenol by Engineered Escherichia coli: Structural Elucidation by Using the In Silico Tools. Molecules 2021, 26, 6264. https://doi.org/10.3390/molecules26206264
Klamrak A, Nabnueangsap J, Puthongking P, Nualkaew N. Synthesis of Ferulenol by Engineered Escherichia coli: Structural Elucidation by Using the In Silico Tools. Molecules. 2021; 26(20):6264. https://doi.org/10.3390/molecules26206264
Chicago/Turabian StyleKlamrak, Anuwatchakij, Jaran Nabnueangsap, Ploenthip Puthongking, and Natsajee Nualkaew. 2021. "Synthesis of Ferulenol by Engineered Escherichia coli: Structural Elucidation by Using the In Silico Tools" Molecules 26, no. 20: 6264. https://doi.org/10.3390/molecules26206264
APA StyleKlamrak, A., Nabnueangsap, J., Puthongking, P., & Nualkaew, N. (2021). Synthesis of Ferulenol by Engineered Escherichia coli: Structural Elucidation by Using the In Silico Tools. Molecules, 26(20), 6264. https://doi.org/10.3390/molecules26206264