Mimotopes for Mycotoxins Diagnosis Based on Random Peptides or Recombinant Antibodies from Phage Library
Abstract
:1. Introduction
2. M13 Phage Display Technology
2.1. The Structure and Life-Cycle of M13 Bacteriophage
2.2. Phage Display Technology
3. Random Peptide Using for Mycotoxins Detection
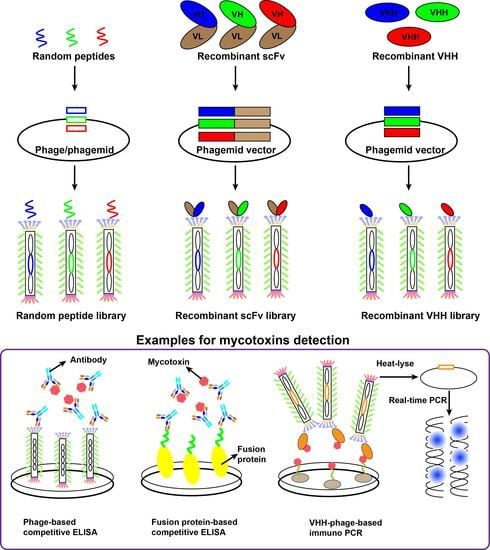
3.1. Random Peptide Libraries
3.2. Random Peptide-Based Mycotoxins Detection
4. Recombinant Antibody Using for Mycotoxins Detection
4.1. Recombinant Antibody Libraries
4.2. ScFv Antbibodies Based-Detection Method
4.3. Anti-Idiotypic Antibody Based-Detection Method
5. Simultaneous Determination of Multiplex Mycotoxins
5.1. Random Peptide-Based Multiplex Detection
5.2. Recombinant Antibodies-Based Multiplex Detection
5.2.1. Time-Resolved Fluorescence Immunochromatographic Assay
5.2.2. Duplex Real-Time PCR Methods
6. Conclusions
Author Contributions
Funding
Conflicts of Interest
References
- Peraica, M.; Radić, B.; Lucić, A.; Pavlović, M. Toxic effects of mycotoxins in humans. Bull. World Health Organ. 1999, 77, 754. [Google Scholar] [PubMed]
- Hussein, H.S.; Brasel, J.M. Toxicity, metabolism, and impact of mycotoxins on humans and animals. Toxicology 2001, 167, 101–134. [Google Scholar] [CrossRef]
- Anfossi, L.; Giovannoli, C.; Baggiani, C. Mycotoxin detection. Curr. Opin. Biotechnol. 2016, 37, 120–126. [Google Scholar] [CrossRef] [PubMed]
- Majdinasab, M.; Ben Aissa, S.; Marty, J.L. Advances in Colorimetric Strategies for Mycotoxins Detection: Toward Rapid Industrial Monitoring. Toxins 2020, 13, 13. [Google Scholar] [CrossRef] [PubMed]
- Chena, F.; Luana, C.; Lin, W.; Shue, W.; Shao, L. Simultaneous determination of six mycotoxins in peanut by high-performance liquid chromatography with a fluorescence detector. J. Sci. Food Agric. 2017, 97, 1805–1810. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Pei, F.; Fang, Y.; Li, P.; Zhao, Y.; Shen, F.; Zou, Y.; Hu, Q. Comparison of concentration and health risks of 9 fusarium mycotoxins in commercial whole wheat flour and refined wheat flour by multi-IAC-HPLC. Food Chem. 2019, 275, 763–769. [Google Scholar] [CrossRef] [PubMed]
- Yelko, R.C.; Juan, C.M.; Houda, B.; Jordi, M. A survey of trichothecenes, zearalenone and patulin in milled grain-based products using GC-MS/MS. Food Chem. 2014, 146, 212–219. [Google Scholar]
- Yelko, R.C.; Juan, C.M.; Jordi, M.; Houda, B. Defvelopment of a GC-MS/MS strategy to determine 15 mycotoxins and metabolites in human urine. Talanta 2014, 128, 125–131. [Google Scholar]
- Kim, E.S.; Chris, R.T.; Stephanie, F.; Martin, H.M.; Michael, E.L.; Chris, M.M.; Lisa, C.S. Indirect competitive immunoassay for detection of aflatoxin B1 in corn and nut products using the array biosensor. Biosens. Bioelectron. 2006, 21, 2298–2305. [Google Scholar]
- Pöhlmann, C.; Elßner, T. Multiplex immunoassay techniques for on-Ssite detection of security sensitive toxins. Toxins 2020, 12, 727. [Google Scholar] [CrossRef] [PubMed]
- Pensuda, S.; Natcha, P.; Kuntalee, R.; Kiattawee, C.; Montarop, Y. Generation of human and rabbit recombinant antibodies for the detection of Zearalenone by phage display antibody technology. Talanta 2019, 201, 397–405. [Google Scholar]
- Wang, J.; Mukhtar, H.; Ma, L.; Pang, Q.; Wang, X.H. VHH antibodies: Reagents for mycotoxin detection in food products. Sensors 2018, 18, 485. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saeed, A.F.; Wang, R.Z.; Ling, S.M.; Wang, S.H. Antibody engineering for pursuing a healthier future. Front. Microbiol. 2017, 8, 495. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jerne, N.K. Towards a network theory of the immune system. Ann. Immunol. (Paris) 1974, 125C, 373–389. [Google Scholar] [PubMed]
- Peltomaa, R.; Barderas, R.; Benito-Peña, E.; Moreno-Bondi, M.C. Recombinant antibodies and their use for food immunoanalysis. Anal. Bioanal. Chem. 2021, 1–25. [Google Scholar] [CrossRef] [PubMed]
- Uhlen, M.; Bandrowski, A.; Carr, S.; Edwards, A.; Ellenberg, J.; Lundberg, E.; Rimm, D.; Rodriguez, H.; Hiltke, T.; Snyder, M.; et al. A proposal for validation of antibodies. Nat. Methods 2016, 13, 823–827. [Google Scholar] [CrossRef] [PubMed]
- Peltomaa, R.; Agudo-Maestro, I.; Más, V.; Barderas, R.; Benito-Peña, E.; Moreno-Bondi, M.C. Development and comparison of mimotope-based immunoassays for the analysis of fumonisin B1. Anal. Bioanal. Chem. 2019, 411, 6801–6811. [Google Scholar] [CrossRef]
- Sheedy, C.; MacKenzie, C.R.; Hall, J.C. Isolation and affinity maturation of hapten-specific antibodies. Biotechnol. Adv. 2007, 25, 333–352. [Google Scholar] [CrossRef]
- Yan, J.X.; Hu, W.J.; You, K.H.; Ma, Z.E.; Xu, Y.; Li, Y.P.; He, Q.H. Biosynthetic mycotoxin conjugate mimetics-mediated green strategy for multiplex mycotoxin immunochromatographic assay. J. Agric. Food Chem. 2020, 68, 2193–2200. [Google Scholar] [CrossRef]
- Shua, M.; Xu, Y.; Wang, D.; Liu, X.; Li, Y.P.; He, Q.H.; Tu, Z.; Qiu, Y.L.; Ji, Y.W.; Wang, X.X. Anti-idiotypic nanobody: A strategy for development of sensitive and green immunoassay for fumonisin B1. Talanta 2015, 143, 388–393. [Google Scholar] [CrossRef]
- He, T.; Zhu, J.; Nie, Y.; Hu, R.; Wang, T.; Li, P.W.; Zhang, Q.; Yang, Y.H. Nanobody technology for mycotoxin detection: Current status and prospects. Toxins 2018, 10, 180. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.J.; McCoy, M.R.; Majkova, Z.; Dechant, J.E.; Gee, S.J.; Tabares-da Rosa, S.; González-Sapienza, G.G.; Hammock, B.D. Isolation of alpaca anti-hapten heavy chain single domain antibodies for development of sensitive immunoassay. Anal. Chem. 2012, 84, 1165–1171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Qiu, Y.L.; He, Q.H.; Xu, Y.; Bhunia, A.K.; Tu, Z.; Chen, B.; Liu, Y.Y. Deoxynivalenol-mimic nanobody isolated from a naive phage display nanobody library and its application in immunoassay. Anal. Chim. Acta 2015, 887, 201–208. [Google Scholar] [CrossRef] [PubMed]
- Arap, W.; Kolonin, M.G.; Trepel, M.; Lahdenranta, J.; Cardó-Vila, M.; Giordano, R.J.; Mintz, P.J.; Ardelt, P.U.; Yao, V.J.; Vidal, C.I.; et al. Steps toward mapping the human vasculature by phage display. Nat. Med. 2002, 8, 121–127. [Google Scholar] [CrossRef] [PubMed]
- Itay, M.; Tomer, S.; Nimrod, D.R.; Jonathan, M.G.; Eytan, R.; Roded, S.; Tal, P. Epitope mapping using combinatorial phage-display libraries: A graph-based algorithm. Nucleic Acids Res. 2007, 35, 69–78. [Google Scholar]
- Cai, J.; Liu, Z.F.; Wang, F.; Li, F. Phage display applications for molecular imaging. Curr. Pharm. Biotechnol. 2010, 11, 603–609. [Google Scholar] [CrossRef]
- Kamada, H.; Okamoto, T.; Kawamura, M.; Shibata, H.; Abe, Y.; Ohkawa, A.; Nomura, T.; Sato, M.; Mukai, Y.; Sugita, T.; et al. Creation of novel cell-penetrating peptides for intracellular drug delivery using systematic phage display technology originated from Tat transduction domain. Biol. Pharm. Bull. 2007, 30, 218–223. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ju, Z.G.; Sun, W. Drug delivery vectors based on filamentous bacteriophages and phage-mimetic nanoparticles. Drug Deliv. 2017, 24, 1898–1908. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sachdev, S.S.; Wayne, J.F.; Kurt, D. Exploring protein-protein interactions with phage display. Chembiochem 2003, 4, 14–25. [Google Scholar]
- Catherine, L.B.; Amos, O.; Andrei, T.; Gali, P.; Sankar, A. A phage display system designed to detect and study protein-protein interactions. Mol. Microbiol. 2008, 67, 719–728. [Google Scholar]
- Yuan, Q.P.; James, J.P.; Brandon, M.H.; Leslie, A.K.; John, E.L.; Hart, L.P. Identification of mimotope peptides which bind to the mycotoxin deoxynivalenol-specific monoclonal antibody. Appl. Environ. Microbiol. 1999, 65, 3279–3286. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zou, X.Q.; Chen, C.C.; Huang, X.L.; Chen, X.L.; Wang, L.; Xiong, Y.H. Phage-free peptide ELISA for ochratoxin A detection based on biotinylated mimotope as a competing antigen. Talanta 2016, 146, 394–400. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, H.; Li, P.; Zhang, Q.; Kim, H.J.; Gee, S.J.; Hammock, B.D. Phage-displayed peptides that mimic aflatoxins and its application in immunoassay. J. Agric. Food Chem. 2013, 61, 2426–2433. [Google Scholar] [CrossRef] [PubMed]
- He, Q.H.; Xu, Y.; Huang, Y.H.; Liu, R.R.; Huang, Z.B.; Li, Y.P. Phage-displayed peptides that mimic zearalenone and its application in immunoassay. Food Chem. 2011, 126, 1312–1315. [Google Scholar] [CrossRef]
- Speijers, G.J.A.; Speijers, M.H.M. Combined toxic effects of mycotoxins. Toxicol. Lett. 2004, 153, 91–98. [Google Scholar] [CrossRef] [PubMed]
- Jong-Sik, M.; Eun, J.C.; Jeong, N.N.; Jong-Ryeul, S.; Han, D.W.; Oh, J.W. Research progress of M13 bacteriophage-based biosensors. Nanomaterials 2019, 9, 1448. [Google Scholar]
- Kuzmicheva, G.A.; Belyavskaya, V.A. Peptide phage display in biotechnology and biomedicine. Biochem. Mosc.-Suppl. S 2017, 11, 1–15. [Google Scholar] [CrossRef]
- Leila, R.; Safar, F.; Hossein, B.; Jafar, M.; Kamal, V.; Vahideh, A.; Bahman, A. Evolution of phage display technology: From discovery to application. J. Drug Target. 2016, 25, 216–224. [Google Scholar]
- Kobra, O.; Maryam, D. Advances in phage display technology for drug discovery. Expert Opin. Drug Dis. 2015, 10, 651–669. [Google Scholar]
- George, P.S. Filamentous fusion phage: Novel expression vectors that display cloned antigens on the virion surface. Science 1985, 228, 1315–1317. [Google Scholar]
- George, P.S.; Valery, A.P. Phage Display. Chem. Rev. 1997, 97, 391–410. [Google Scholar]
- Il’ichev, A.A.; Minenkova, O.O.; Tat’kov, S.I.; Karpyshev, N.N.; Eroshkin, A.M.; Petrenko, V.A.; Sandakhchiev, L.S. Production of a viable variant of the M13 phage with a foreign peptide inserted into the basic coat protein. Dokl. Akad. Nauk. SSSR 1989, 307, 481–483. [Google Scholar]
- Jespers, L.S.; Messens, J.H.; De, K.A.; Eeckhout, D.; Brande, I.; Gansemans, Y.G.; Lauwereys, M.J.; Stanssens, P.E. Surface expression and ligand-based selection of cDNAs fused to filamentous phage gene VI. Biotechnology 1995, 13, 378–382. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.C.; Ju, Z.G.; Cao, B.R.; Gao, X.; Zhu, Y.; Qiu, P.H.; Xu, H.; Pan, P.T.; Bao, H.Z.; Wang, L.; et al. Ultrasensitive rapid detection of human serum antibody biomarkers by biomarker-capturing viral nanofibers. ACS Nano 2015, 9, 4475–4483. [Google Scholar] [CrossRef] [Green Version]
- Devlin, J.J.; Panganiban, L.C.; Devlin, P.E. Random peptide libraries: A source of specific protein binding molecules. Science 1990, 249, 404–406. [Google Scholar] [CrossRef] [PubMed]
- Hennie, R.H.; Adriaan, P.D.; Simon, E.H.; René, M.H.; Arends, J.W.; Roovers, R.C. Antibody phage display technology and its applications. Immunotechnology 1998, 4, 1–20. [Google Scholar]
- Mark, R.H.; Luciano, M.; Arthur, P. Tunable alignment of macromolecules by filamentous phage yields dipolar coupling interactions. Nat. Struct. Biol. 1998, 5, 1065–1074. [Google Scholar]
- Scott, J.K.; Smith, G.P. Searching for peptide ligands with an epitope library. Science 1990, 249, 386–390. [Google Scholar] [CrossRef]
- Kehoe, J.W.; Kay, B.K. Filamentous phage display in the new millennium. Chem. Rev. 2005, 105, 4056–4072. [Google Scholar] [CrossRef] [PubMed]
- Sharon, L.B.; Francis, J.S.; Alice, L.E.; Arnold, L.S. Peptides selected for binding to a virulent strain of Haemophilus influenzae by phage display are bactericidal. Antimicrob. Agents Chemother. 2005, 49, 2972–2978. [Google Scholar]
- James, W.G.; Amanda, L.G.; Anatoliy, T.P.; Deepa, B.; Valery, A.P. Combinatorial synthesis and screening of cancer cell-specific nanomedicines targeted via phage fusion proteins. Front. Microbiol. 2015, 6, 628. [Google Scholar]
- Mikhail, G.K.; Sun, J.; Kim-Anh, D.; Claudia, I.V.; Yuan, J.; Keith, A.B.; Renata, P.; Wadih, A. Synchronous selection of homing peptides for multiple tissues by in vivo phage display. FASEB J. 2006, 20, 979–981. [Google Scholar]
- Renata, P.; Erkki, R. Organ targeting In vivo using phage display peptide libraries. Nature 1996, 380, 364–366. [Google Scholar]
- Saggy, I.; Yariv, W.; Leeron, S.C.; Limor, N.; George, G.; Itai, B. Antibody isolation from immunized animals: Comparison of phage display and antibody discovery via V gene repertoire mining. Protein Eng. Des. Sel. 2012, 25, 539–549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Andrew, C.; Bergquist, P.L.; Anwar, S. Solid-binding peptides in biomedicine. Adv. Exp. Med. Biol. 2017, 1030, 21–36. [Google Scholar]
- He, Z.Y.; He, Q.H.; Xu, Y.; Li, Y.P.; Liu, X.; Chen, B.; Lei, D.; Sun, C.H. Ochratoxin A mimotope from second-generation peptide library and its application in immunoassay. Anal. Chem. 2013, 85, 10304–10311. [Google Scholar] [CrossRef]
- Thirumala-Devi, K.; Miller, J.S.; Reddy, G.; Reddy, D.V.R.; Mayo, M.A. Phage-displayed peptides that mimic aflatoxin B1 in serological reactivity. J. Appl. Microbiol. 2001, 90, 330–336. [Google Scholar] [CrossRef] [Green Version]
- Liu, R.R.; Xu, L.; Qiu, X.M.; Chen, X.L.; Deng, H.L.; Lai, W.H.; Xu, Y. An immunoassay for determining aflatoxin B1 using a recombinant phage as a nontoxic coating conjugate. J. Food Saf. 2012, 32, 318–325. [Google Scholar] [CrossRef]
- Liu, R.R.; Yu, Z.; He, Q.H.; Wang, X.; Xu, Y. Selecting mimotope of ochratoxin A from phage random peptide library and its application. Chin. J. Public Health 2005, 21, 844–946. [Google Scholar]
- He, Q.H.; Xu, Y.; Zhang, C.Z.; Li, Y.P.; Huang, Z.B. Phage-borne peptidomimetics as immunochemical reagent in dot-immunoassay for mycotoxin zearalenone. Food Control 2014, 39, 56–61. [Google Scholar] [CrossRef]
- Liu, X.; Xu, Y.; He, Q.H.; He, Z.Y.; Xiong, Z.P. Application of mimotope peptides of fumonisin B1 in peptide ELISA. J. Agric. Food Chem. 2013, 61, 4765–4770. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Chen, B.; He, Q.H.; Qiu, Y.L.; Liu, X.; He, Z.Y.; Xiong, Z.P. New approach for development of sensitive and environmentally friendly immunoassay for mycotoxin fumonisin B1 based on using peptide-MBP fusion protein as substitute for coating antigen. Anal. Chem. 2014, 86, 8433–8440. [Google Scholar] [CrossRef] [PubMed]
- Riikka, P.; Elena, B.P.; Rodrigo, B.; Ursula, S.; Martin, G.A.; María, C.M.B. Microarray-based immunoassay with synthetic mimotopes for the detection of fumonisin B1. Anal. Chem. 2017, 89, 6216–6223. [Google Scholar]
- Yu, M.; Than, K.; Colegate, S.; Shiell, B.; Wojtek, P.M.; Stephen, P.; Wang, L.F. Peptide mimotopes of phomopsins: Identification, characterization and application in an immunoassay. Mol. Divers. 2005, 9, 233–240. [Google Scholar] [CrossRef] [PubMed]
- Nitsara, K.; Ratthaphol, C.; Morton, M.; Michalina, O.S.; Irene, R.G.; Christopher, T.E. Development of a M13 bacteriophage-based SPR detection using Salmonella as a case study. Sens. Actuators B-Chem. 2014, 190, 214–220. [Google Scholar]
- Yang, S.L.; Shang, Y.L.; Yin, S.H.; Wang, D.; Cai, J.P.; Gong, Z.L.; Serge, M.; Liu, X.T. A phage-displayed single domain antibody fused to alkaline phosphatase for detection of porcine circovirus type 2. J. Virol. Methods 2015, 213, 84–92. [Google Scholar] [CrossRef] [PubMed]
- Kuang, Z.F.; Sang, N.K.; Wendy, J.C.; Barry, L.F.; Rajesh, R.N. Biomimetic chemosensor: Designing peptide recognition elements for surface functionalization of carbon nanotube field effect transistors. ACS Nano 2010, 4, 452–458. [Google Scholar] [CrossRef]
- Cristiano, C.; Jan, W.V.; Richard, M.T.; Pier, L.L. Investigation of de novo yotally random biosequences, part I: A general method for in vitro selection of folded domains from a random polypeptide library displayed on phage. Chem. Biodivers. 2006, 3, 827–839. [Google Scholar]
- Guo, J.; Jeffrey, M.C.; Mohamed, N.A.M.; Benesi, A.J.; Tien, M.; Kao, T.H.; Watts, H.D.; Kubicki, J.D. Identification and characterization of a cellulose binding heptapeptide revealed by phage display. Biomacromolecules 2013, 14, 1795–1805. [Google Scholar] [CrossRef] [PubMed]
- Liu, F.F.; Wang, T.; Dong, X.Y.; Sun, Y. Rational design of affinity peptide ligand by flexible docking simulation. J. Chromatogr. A 2007, 1146, 41–50. [Google Scholar] [CrossRef]
- Lai, W.H.; Daniel, Y.C.F.; Xu, Y.; Liu, R.R.; Xiong, Y.H. Development of a colloidal gold strip for rapid detection of ochratoxin A with mimotope peptide. Food Control 2009, 20, 791–795. [Google Scholar] [CrossRef]
- Xue, S.; Li, H.P.; Zhang, J.B.; Liu, J.L.; Hu, Z.Q.; Gong, A.D.; Huang, T.; Liao, Y.C. Chicken single-chain antibody fused to alkaline phosphatase detects Aspergillus pathogens and their presence in natural samples by direct sandwich enzyme-linked immunosorbent assay. Anal. Chem. 2013, 85, 10992–10999. [Google Scholar] [CrossRef] [PubMed]
- Viti, F.; Nilsson, F.; Demartis, S.; Huber, A.; Neri, D. Design and use of phage display libraries for the selection of antibodies and enzymes. Methods Enzymol. 2000, 326, 480–505. [Google Scholar] [PubMed]
- Yang, L.; Ding, H.; Gu, Z.; Zhao, J.; Chen, H.; Tian, F.; Chen, Y.Q.; Zhang, H.; Chen, W. Selection of single chain fragment variables with direct coating of aflatoxin B1 to enzyme-linked immunosorbent assay plates. J. Agric. Food Chem. 2009, 57, 8927–8932. [Google Scholar] [CrossRef]
- Xu, Y.; Xiong, L.; Li, Y.; Xiong, Y.; Tu, Z.; Fu, J.; Chen, B. Anti-idiotypic nanobody as citrinin mimotope from a naive alpaca heavy chain single domain antibody library. Anal. Bioanal. Chem. 2015, 407, 5333–5341. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; He, Q.; Xu, Y.; Liu, X.; Shu, M.; Tu, Z.; Li, Y.; Wang, W.; Cao, D. Anti-idiotypic VHH phage display-mediated immuno-PCR for ultrasensitive determination of mycotoxin zearalenone in cereals. Talanta 2016, 147, 410–415. [Google Scholar] [CrossRef]
- Tu, Z.; Xu, Y.; He, Q.H.; Fu, J.H.; Liu, X.; Tao, Y. Isolation and characterisation of deoxynivalenol affinity binders from a phage display library based on single-domain camelid heavy chain antibodies (VHHs). Food Agric. Immunol. 2012, 23, 123–131. [Google Scholar] [CrossRef]
- Wang, Y.; Li, P.; Majkova, Z.; Bever, C.R.; Kim, H.J.; Zhang, Q.; Dechant, J.E.; Gee, S.J.; Hammock, B.D. Isolation of alpaca anti-idiotypic heavy-chain single-domain antibody for the aflatoxin immunoassay. Anal. Chem. 2013, 85, 8298–8303. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bird, R.E.; Hardman, K.D.; Jacobson, J.W.; Johnson, S.; Kaufman, B.M.; Lee, S.M.; Lee, T.; Pope, S.H.; Riordan, G.S.; Whitlow, M. Single-chain antigen-binding proteins. Science 1988, 242, 423–426. [Google Scholar] [CrossRef] [PubMed]
- Huston, J.S.; Levinson, D.; Mudgett-Hunter, M.; Tai, M.S.; Novotný, J.; Margolies, M.N.; Ridge, R.J.; Bruccoleri, R.E.; Haber, E.; Crea, R. Protein engineering of antibody binding sites: Recovery of specific activity in an anti-digoxin single-chain Fv analogue produced in Escherichia coli. Proc. Natl. Acad. Sci. USA 1988, 85, 5879–5883. [Google Scholar] [CrossRef] [Green Version]
- Bemani, P.; Mohammadi, M.; Hakakian, A. ScFv Improvement approaches. Protein Pept. Lett. 2018, 25, 222–229. [Google Scholar] [CrossRef] [PubMed]
- Zou, L.; Xu, Y.; Li, Y.; He, Q.; Chen, B.; Wang, D. Development of a single-chain variable fragment antibody-based enzyme-linked immunosorbent assay for determination of fumonisin B₁ in corn samples. J. Sci. Food Agric. 2014, 94, 1865–1871. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Q.; Clarke, J.R.; Zhou, H.R.; Linz, J.E.; Pestka, J.J.; Hart, L.P. Molecular cloning, expression, and characterization of a functional single-chain Fv antibody to the mycotoxin zearalenone. Appl. Environ. Microbiol. 1997, 63, 263–269. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.H.; Du, X.Y.; Huang, Y.M.; Lin, D.S.; Hart, P.L.; Wang, Z.H. Detection of deoxynivalenol based on a single-chain fragment variable of the antideoxynivalenol antibody. FEMS Microbiol. Lett. 2007, 272, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Gu, X.; Zhuang, Z.; Zhong, Y.; Yang, H.; Wang, S. Screening and Molecular Evolution of a Single Chain Variable Fragment Antibody (scFv) against Citreoviridin Toxin. J. Agric. Food Chem. 2016, 64, 7640–7648. [Google Scholar] [CrossRef]
- Rangnoi, K.; Jaruseranee, N.; O’Kennedy, R.; Pansri, P.; Yamabhai, M. One-step detection of aflatoxin-B(1) using scFv-alkaline phosphatase-fusion selected from human phage display antibody library. Mol. Biotechnol. 2011, 49, 240–249. [Google Scholar] [CrossRef] [PubMed]
- Moghaddam, A.; Løbersli, I.; Gebhardt, K.; Braunagel, M.; Marvik, O.J. Selection and characterisation of recombinant single-chain antibodies to the hapten aflatoxin-B1 from naive recombinant antibody libraries. J. Immunol. Methods 2001, 254, 169–181. [Google Scholar] [CrossRef]
- Li, X.; Li, P.W.; Lei, J.W.; Zhang, Q.; Zhang, W.; Li, C.M. A simple strategy to obtain ultra-sensitive single-chain fragment variable antibodies for aflatoxin detection. RSC Adv. 2013, 3, 22367. [Google Scholar] [CrossRef]
- Rangnoi, K.; Choowongkomon, K.; O’Kennedy, R.; Rüker, F.; Yamabhai, M. Enhancement and analysis of human antiaflatoxin B1 (AFB1) scFv antibody-ligand interaction using chain shuffling. J. Agric. Food Chem. 2018, 66, 5713–5722. [Google Scholar] [CrossRef]
- Zhang, C.; Zhang, Q.; Tang, X.; Zhang, W.; Li, P. Development of an Anti-Idiotypic VHH Antibody and Toxin-Free Enzyme Immunoassay for Ochratoxin A in Cereals. Toxins 2019, 11, 280. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Xu, Y.; Xiong, Y.H.; Tu, Z.; Li, Y.P.; He, Z.Y.; Qiu, Y.L.; Fu, J.H.; Gee, S.J.; Hammock, B.D. VHH phage-based competitive real-time immuno-polymerase chain reaction for ultrasensitive detection of ochratoxin A in cereal. Anal. Chem. 2014, 86, 7471–7477. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Xu, Y.; Wan, D.B.; Xiong, Y.H.; He, Z.Y.; Wang, X.X.; Gee, S.J.; Ryu, D.; Hammock, B.D. Development of a nanobody-alkaline phosphatase fusion protein and its application in a highly sensitive direct competitive fluorescence enzyme immunoassay for detection of ochratoxin A in cereal. Anal. Chem. 2015, 87, 1387–1394. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Doyle, P.J.; Arbabi-Ghahroudi, M.; Gaudette, N.; Furzer, G.; Savard, M.E.; Gleddie, S.; McLean, M.D.; Mackenzie, C.R.; Hall, J.C. Cloning, expression, and characterization of a single-domain antibody fragment with affinity for 15-acetyl-deoxynivalenol. Mol. Immunol. 2008, 45, 3703–3713. [Google Scholar] [CrossRef] [PubMed]
- Yu, F.Y.; Chu, F.S. Production and characterization of a monoclonal anti-anti-idiotype antibody against fumonisin B1. J. Agric. Food Chem. 1999, 47, 4815–4820. [Google Scholar] [CrossRef] [PubMed]
- Guan, D.; Li, P.; Cui, Y.; Zhang, Q.; Zhang, W. A competitive immunoassay with a surrogate calibrator curve for aflatoxin M1 in milk. Anal. Chim. Acta 2011, 703, 64–69. [Google Scholar] [CrossRef] [PubMed]
- Hamers-Casterman, C.; Atarhouch, T.; Muyldermans, S.; Robinson, G.; Hamers, C.; Songa, E.B.; Bendahman, N.; Hamers, R. Naturally occurring antibodies devoid of light chains. Nature 1993, 363, 446–448. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S.; Atarhouch, T.; Saldanha, J.; Barbosa, J.A.; Hamers, R. Sequence and structure of VH domain from naturally occurring camel heavy chain immunoglobulins lacking light chains. Protein Eng. 1994, 7, 1129–1135. [Google Scholar] [CrossRef] [PubMed]
- Muyldermans, S.; Baral, T.N.; Retamozzo, V.C.; De Baetselier, P.; De Genst, E.; Kinne, J.; Leonhardt, H.; Magez, S.; Nguyen, V.K.; Revets, H.; et al. Camelid immunoglobulins and nanobody technology. Vet. Immunol. Immunopathol. 2009, 128, 178–183. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zarebski, L.M.; Urrutia, M.; Goldbaum, F.A. Llama single domain antibodies as a tool for molecular mimicry. J. Mol. Biol. 2005, 349, 814–824. [Google Scholar] [CrossRef]
- Tanha, J.; Dubuc, G.; Hirama, T.; Narang, S.A.; MacKenzie, C.R. Selection by phage display of llama conventional V(H) fragments with heavy chain antibody V(H)H properties. J. Immunol. Methods 2002, 263, 97–109. [Google Scholar] [CrossRef]
- Guo, Y.C.; Zhou, Y.F.; Zhang, X.E.; Zhang, Z.P.; Qiao, Y.M.; Bi, L.J.; Wen, J.K.; Liang, M.F.; Zhang, J.B. Phage display mediated immuno-PCR. Nucleic Acids Res. 2006, 34, e62. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kim, H.J.; Mark, M.C.; Shirley, J.G.; Gualberto, G.G.; Bruce, D.H. Noncompetitive phage anti-immunocomplex real-time polymerase chain reaction for sensitive detection of small molecules. Anal. Chem. 2011, 83, 246–253. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dong, J.H.; Hasan, S.; Yuuichiro, F.; Hiroshi, U. Detection of small molecule diagnostic markers with phage-based open-sandwich immuno-PCR. J. Immunol. Methods 2012, 377, 1–7. [Google Scholar] [CrossRef] [PubMed]
- Notomi, T.; Okayama, H.; Masubuchi, H.; Yonekawa, T.; Watanabe, K.; Amino, N.; Hase, T. Loop-mediated isothermal amplification of DNA. Nucleic Acids Res. 2000, 28, e63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sagcan, H.; Kara, N.T. Detection of Potato ring rot Pathogen Clavibacter michiganensis subsp. s epedonicus by Loop-mediated isothermal amplification (LAMP) assay. Sci. Rep. 2019, 9, 20393. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rivero, R.; Bisi, M.; Velázquez, E.B.; Esteva, M.I.; Scollo, K.; González, N.L.; Altcheh, J.; Ruiz, A.M. Rapid detection of Trypanosoma cruzi by colorimetric loop-mediated isothermal amplification (LAMP): A potential novel tool for the detection of congenital Chagas infection. Diagn. Microbiol. Infect. Dis. 2017, 89, 26–28. [Google Scholar] [CrossRef] [PubMed]
- Lei, J.; Han, X.; Tang, X.; Wang, H.; Zhang, Q. Development of Anti-Idiotypic Nanobody-Phage Based Immuno-Loop-Mediated Isothermal Amplification Assay for Aflatoxins in Peanuts. Toxins 2020, 12, 565. [Google Scholar] [CrossRef] [PubMed]
- Vinay, V.; Michael, S.; Roman, L.; Wolfgang, B.; Rudolf, K. Simultaneous determination of 186 fungal and bacterial metabolites in indoor matrices by liquid chromatography/tandem mass spectrometry. Anal. Bioanal. Chem. 2009, 395, 1355–1372. [Google Scholar]
- Pamel, E.V.; Annemieke, V.; Geertrui, V.; Johan, D.B.; Els, D. Ultrahigh-performance liquid chromatographic-tandem mass spectrometric multimycotoxin method for quantitating 26 mycotoxins in maize silage. J. Agric. Food Chem. 2011, 59, 9747–9755. [Google Scholar] [CrossRef] [PubMed]
- Debjani, S.; Debopam, A.; Dipika, R.; Dilip, S.; Tarun, K.D. Simultaneous enzyme immunoassay for the screening of aflatoxin B1 and ochratoxin A in chili samples. Anal. Chim. Acta 2007, 584, 343–349. [Google Scholar]
- Wu, S.J.; Duan, N.; Ma, X.Y.; Xia, Y.; Wang, H.X.; Wang, Z.P.; Zhang, Q. Multiplexed fluorescence resonance energy transfer aptasensor between upconversion nanoparticles and graphene oxide for the simultaneous determination of mycotoxins. Anal. Chem. 2012, 84, 6263–6270. [Google Scholar] [CrossRef]
- Xing, C.; Dong, X.; Xu, T.; Yuan, J.; Yan, W.; Sui, X.; Zhao, X. Analysis of multiple mycotoxins-contaminated wheat by a smart analysis platform. Anal. Biochem. 2020, 610, 113928. [Google Scholar] [CrossRef] [PubMed]
- Xu, L.; Zhang, Z.; Zhang, Q.; Zhang, W.; Yu, L.; Wang, D.; Li, H.; Li, P. An on-site simultaneous semi-quantification of aflatoxin B1, zearalenone, and T-2 toxin in maize- and cereal-based feed via multicolor immunochromatographic assay. Toxins 2018, 10, 87. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Chen, Q.; Han, M.; Zhou, J.; Gong, L.; Niu, Y.; Zhang, Y.; He, L.; Zhang, L. Development and optimization of a multiplex lateral flow immunoassay for the simultaneous determination of three mycotoxins in corn, rice and peanut. Food Chem. 2016, 213, 478–484. [Google Scholar] [CrossRef] [PubMed]
- Li, R.; Meng, C.; Wen, Y.; Fu, W.; He, P. Fluorometric lateral flow immunoassay for simultaneous determination of three mycotoxins (aflatoxin B1, zearalenone and deoxynivalenol) using quantum dot microbeads. Mikrochim. Acta 2019, 186, 748. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.; Yu, X.; Wen, K.; Li, C.; Mujtaba Mari, G.; Jiang, H.; Shi, W.; Shen, J.; Wang, Z. Multiplex lateral flow immunoassays based on amorphous carbon nanoparticles for detecting three fusarium mycotoxins in maize. J. Agric. Food Chem. 2017, 65, 8063–8071. [Google Scholar] [CrossRef] [PubMed]
- Lattanzio, V.M.T.; Guarducci, N.; Powers, S.; Ciasca, B.; Pascale, M.; von Holst, C. Validation of a lateral flow immunoassay for the rapid determination of aflatoxins in maize by solvent free extraction. Anal. Methods 2017, 10, 123–130. [Google Scholar] [CrossRef]
- Butler, S.A.; Khanlian, S.A.; Cole, L.A. Detection of early pregnancy forms of human chorionic gonadotropin by home pregnancy test devices. Clin. Chem. 2001, 47, 2131–2136. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, J.; Lee, S.; Choo, J. Application of a SERS-based lateral flow immunoassay strip for the rapid and sensitive detection of staphylococcal enterotoxin B. Nanoscale 2016, 8, 11418–11425. [Google Scholar] [CrossRef] [PubMed]
- Anfossi, L.; Di Nardo, F.; Cavalera, S.; Giovannoli, C.; Baggiani, C. Multiplex lateral flow immunoassay: An overview of strategies towards high-throughput point-of-need testing. Biosensors 2018, 9, 2. [Google Scholar] [CrossRef] [Green Version]
- Jawaid, W.; Campbell, K.; Melville, K.; Holmes, S.J.; Rice, J.; Elliott, C.T. Development and Validation of a Novel Lateral Flow Immunoassay (LFIA) for the Rapid Screening of Paralytic Shellfish Toxins (PSTs) from Shellfish Extracts. Anal. Chem. 2015, 87, 5324–5332. [Google Scholar] [CrossRef] [PubMed]
- Wang, C.; Xiao, R.; Wang, S.; Yang, X.; Bai, Z.; Li, X.; Rong, Z.; Shen, B.; Wang, S. Magnetic quantum dot based lateral flow assay biosensor for multiplex and sensitive detection of protein toxins in food samples. Biosens. Bioelectron. 2019, 146, 111754. [Google Scholar] [CrossRef]
- Climent, E.; Biyikal, M.; Gröninger, D.; Weller, M.G.; Martínez-Máñez, R.; Rurack, K. Multiplexed detection of analytes on single test strips with antibody-gated indicator-releasing mesoporous nanoparticles. Angew. Chem. Int. Ed. 2020, 59, 23862–23869. [Google Scholar] [CrossRef] [PubMed]
- Tang, X.; Li, P.; Zhang, Q.; Zhang, Z.; Zhang, W.; Jiang, J. Time-Resolved Fluorescence Immunochromatographic Assay Developed Using Two Idiotypic Nanobodies for Rapid, Quantitative, and Simultaneous Detection of Aflatoxin and Zearalenone in Maize and Its Products. Anal. Chem. 2017, 89, 11520–11528. [Google Scholar] [CrossRef] [PubMed]
- Ren, X.; Yue, X.; Mwakinyali, S.E.; Zhang, W.; Zhang, Q.; Li, P. Small Molecular Contaminant and Microorganism Can Be Simultaneously Detected Based on Nanobody-Phage: Using Carcinogen Aflatoxin and Its Main Fungal Aspergillus Section Flavi spp. in Stored Maize for Demonstration. Front. Microbiol. 2020, 10, 3023. [Google Scholar] [CrossRef] [PubMed]
- Shkembi, X.; Svobodova, M.; Skouridou, V.; Bashammakh, A.S.; Alyoubi, A.O.; O’Sullivan, C.K. Aptasensors for mycotoxin detection: A review. Anal. Biochem. 2021, in press. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.Y.; Hsiao, J.K.; Wang, Y.P.; Lan, C.H.; Wu, H.C. Peptide-conjugated nanoparticles for targeted imaging and therapy of prostate cancer. Biomaterials 2016, 99, 1–15. [Google Scholar] [CrossRef] [PubMed]





| Mycotoxin | Library | Passenger Protein | Peptide Mimics | Linear Range | LOD | Detection Principle | Sample Matrices | Ref. |
|---|---|---|---|---|---|---|---|---|
| DON | Random 7-mer peptide library | pIII | SWGPFPF; SWGPLPF | 0.1–10 µg/mL | - | Competitive ELISA | Wheat | [31] |
| AFB1 | Random 8-mer peptide library | pIII | -PHPWNP-; -T-HRNW- | 4–24 µg/kg | - | Competitive ELISA | Peanut and feedstuff | [33] |
| Random Cys-4/Cys-6 peptide library | pVIII | CYMD-C | - | - | Competitive ELISA | Groundnut | [57] | |
| Ph.D.™-7 Phage Display Peptide Library | pIII | HPSDPRH | 100–2500 pg/mL | - | Competitive ELISA | Rice, wheat, corn, and feedstuff | [58] | |
| OTA | Random 7-mer peptide library | pIII | GMVQTIF | 0.005–0.2 ng/mL | 0.1 ng/mL | Competitive ELISA | Corn | [32] |
| Second generation peptide library | pIII | AETYGFQLHAMK | 0.006–0.245 ng/mL | 0.005 ng/mL | Chemiluminescent ELISA | Corn, rice, and instant coffee | [56] | |
| Ph.D.™-7 phage display peptide library | pIII | IRPMVXX | 200–8000 pg/mL | 150 pg/mL | Competitive ELISA | - | [59] | |
| ZEN | Ph.D.™-7 Phage Display Peptide Library | pIII | DAVILLM; HHCHWWH | 100–10,000 pg/mL | 100 pg/mL | Competitive ELISA | Wheat, corn, and feedstuff | [34] |
| Random 12-mer peptide library | pIII | ESYWATVPWTRH | 50–100 µg/kg | - | Dot-immunoassay | Peanut, corn and rice | [60] | |
| FB1 | Ph.D.-C7C phage display peptide library | pIII | E-L-P-T-L | 1.77–20.73 ng/mL | 1.18 ng/mL | Chemiluminescent Immunoassay | Maize, feedstuff, and wheat | [61] |
| Random 12-mer peptide library | pIII | NNAAMYSEMATD; TTLQMRSEMADD | - | 0.21 ng/mL | Elispot Immunoassay | Maize, feedstuff, and rice | [62] | |
| Ph.D.™-12 Phage Display Peptide Library | pIII | VTPNDDTFDPFR | 17.3–79.6 ng/mL | 11.1 ng/mL | Microarray-based Immunoassay | Maize and wheat | [63] | |
| Phomopsin | Random 15-mer phage display peptide library | pIII | CTVALCNMYFGAKLD | - | - | Competitive ELISA | Lupin seed | [64] |
| Mycotoxin | Antibody Type | Library | Linear Range | LOD | IC50 | Detection Principle | Sample Matrices | Ref. |
|---|---|---|---|---|---|---|---|---|
| FB1 | scFv | VHH library | 2.10–76.45 µg/L | 8.32 µg/kg | 12.67 µg/L | Competitive ELISA | Corn | [82] |
| ZEN | scFv | VHH library | - | - | 17 ng/mL | Competitive ELISA | Corn | [83] |
| DON | scFv | - | - | - | 8.2 ± 0.6 ng/mL | Competitive ELISA | Wheat | [84] |
| CIT | scFv | Mutational phage library | 25–562 µg/mL | 14.7 ng/mL | 120 ng/mL | Competitive ELISA | Corn | [85] |
| AFB1 | scFv | Human non-immunized scFv library | 0.007–0.2 µg/mL | 0.007 µg/mL | 0.034 µg/mL | Competitive ELISA | - | [86] |
| AFB1 | scFv | Naive recombinant antibody libraries | - | - | Competitive ELISA | - | [87] | |
| AFB1 | scFv | Tomlinson libraries I + J | 0.4 ng/mL | Competitive ELISA | [74] | |||
| AFB1 | scFv | positive phage-display library | - | - | 0.01 ng/mL | Competitive ELISA | - | [88] |
| AFB1 | scFv | Variable VH/VL shuffled library | 0.019–5 µg/mL | 0.02 µg/mL | Competitive ELISA | - | [89] | |
| CIT | VHH | Naive alpaca phage displayed VHH library | 5–300 ng/mL | 7.6 μg/kg; 8.6 μg/kg | 44.6 ng/mL | VHH-based ELISA | Wheat, Rice | [75] |
| OTA | VHH | - | 0.003–0.673 ng/mL | 0.001 ng/mL | 0.097 µg/mL | Competitive ELISA | Corn, rice, wheat | [90] |
| OTA | VHH | VHH Library | 0.01–1000 pg/mL | 3.7 pg/L, | 0.31 ng/mL | PD-IPCR | Corn, wheat, rice | [91] |
| OTA | VHH | - | 0.06–0.43 ng/mL | 0.04 ng/mL | 0.13 ng/mL | Fluorescencecompetitive ELISA | Rice, oats, barley | [92] |
| 15-AcDON | VHH | VHH library | 10–5000 ng/mL | 19 ng/mL | 0.5 µM | Competitive ELISA | - | [93] |
| ZEN | VHH | Naive alpaca phage displayed VHH library | 0.11–0.55 ng/mL | 0.08 ng/mL | 0.25 ± 0.02 ng/mL | PD-IPCR | Corn, wheat, rice | [76] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sun, W.; Zhang, Y.; Ju, Z. Mimotopes for Mycotoxins Diagnosis Based on Random Peptides or Recombinant Antibodies from Phage Library. Molecules 2021, 26, 7652. https://doi.org/10.3390/molecules26247652
Sun W, Zhang Y, Ju Z. Mimotopes for Mycotoxins Diagnosis Based on Random Peptides or Recombinant Antibodies from Phage Library. Molecules. 2021; 26(24):7652. https://doi.org/10.3390/molecules26247652
Chicago/Turabian StyleSun, Wei, Yan Zhang, and Zhigang Ju. 2021. "Mimotopes for Mycotoxins Diagnosis Based on Random Peptides or Recombinant Antibodies from Phage Library" Molecules 26, no. 24: 7652. https://doi.org/10.3390/molecules26247652
APA StyleSun, W., Zhang, Y., & Ju, Z. (2021). Mimotopes for Mycotoxins Diagnosis Based on Random Peptides or Recombinant Antibodies from Phage Library. Molecules, 26(24), 7652. https://doi.org/10.3390/molecules26247652