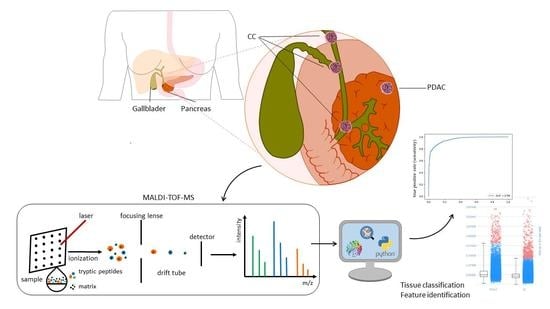
MALDI Mass Spectrometry Imaging for the Distinction of Adenocarcinomas of the Pancreas and Biliary Tree
Abstract
:1. Introduction
2. Materials and Methods
3. Results
3.1. Patient Characteristics
3.2. Supervised Classification
3.3. Discriminative m/z Values
- m/z 3086.53, 3085.52, 850.43, 3101.51, 2104.07, 2073.05, 3102.53, 2056.07, 1199.67, 3100.51, for calculation on the whole dataset
- m/z 2692.33, 3086.53, 1138.55, 1199.67, 1460.70, 816.38, 3085.52, 850.43, 2073.05, 1581.75 for calculation on the reduced dataset
3.4. Tentative m/z Identification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Sample Availability
References
- Kamisawa, T.; Wood, L.D.; Itoi, T.; Takaori, K. Pancreatic cancer. Lancet 2016, 388, 73–85. [Google Scholar] [CrossRef]
- Valle, J.W.; Borbath, I.; Khan, S.A.; Huguet, F.; Gruenberger, T.; Arnold, D.; Committee, E.G. Biliary cancer: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol. 2016, 27, v28–v37. [Google Scholar] [CrossRef] [PubMed]
- Nagtegaal, I.D.; Odze, R.D.; Klimstra, D.; Paradis, V.; Rugge, M.; Schirmacher, P.; Washington, K.M.; Carneiro, F.; Cree, I.A.; WHO Classification of Tumours Editorial Board. The 2019 WHO classification of tumours of the digestive system. Histopathology 2020, 76, 182–188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Cancer-Our World in Data. Available online: https://ourworldindata.org/grapher/total-cancer-deaths-by-type?country=~OWID_WRL (accessed on 14 August 2021).
- Ethun, C.G.; Lopez-Aguiar, A.G.; Pawlik, T.M.; Poultsides, G.; Idrees, K.; Fields, R.C.; Weber, S.M.; Cho, C.; Martin, R.C.; Scoggins, C.R.; et al. Distal Cholangiocarcinoma and Pancreas Adenocarcinoma: Are They Really the Same Disease? A 13-Institution Study from the US Extrahepatic Biliary Malignancy Consortium and the Central Pancreas Consortium. J. Am. Coll. Surg. 2017, 224, 406–413. [Google Scholar] [CrossRef] [PubMed]
- Gonzalez, R.S.; Bagci, P.; Basturk, O.; Reid, M.D.; Balci, S.; Knight, J.H.; Kong, S.Y.; Memis, B.; Jang, K.T.; Ohike, N.; et al. Intrapancreatic distal common bile duct carcinoma: Analysis, staging considerations, and comparison with pancreatic ductal and ampullary adenocarcinomas. Mod. Pathol. 2016, 29, 1358–1369. [Google Scholar] [CrossRef]
- Lee, E.S.; Lee, J.M. Imaging diagnosis of pancreatic cancer: A state-of-the-art review. World J. Gastroenterol. 2014, 20, 7864–7877. [Google Scholar] [CrossRef]
- McKinney, K.Q.; Lee, J.G.; Sindram, D.; Russo, M.W.; Han, D.K.; Bonkovsky, H.L.; Hwang, S.I. Identification of differentially expressed proteins from primary versus metastatic pancreatic cancer cells using subcellular proteomics. Cancer Genom. Proteom. 2012, 9, 257–263. [Google Scholar]
- Le Faouder, J.; Laouirem, S.; Alexandrov, T.; Ben-Harzallah, S.; Leger, T.; Albuquerque, M.; Bedossa, P.; Paradis, V. Tumoral heterogeneity of hepatic cholangiocarcinomas revealed by MALDI imaging mass spectrometry. Proteomics 2014, 14, 965–972. [Google Scholar] [CrossRef]
- Ito, T.; Sakurai-Yageta, M.; Goto, A.; Pairojkul, C.; Yongvanit, P.; Murakami, Y. Genomic and transcriptional alterations of cholangiocarcinoma. J. Hepatobiliary Pancreat. Sci. 2014, 21, 380–387. [Google Scholar] [CrossRef] [Green Version]
- Schwamborn, K.; Caprioli, R.M. Molecular imaging by mass spectrometry—Looking beyond classical histology. Nat. Rev. Cancer 2010, 10, 639–646. [Google Scholar] [CrossRef]
- Porta Siegel, T.; Hamm, G.; Bunch, J.; Cappell, J.; Fletcher, J.S.; Schwamborn, K. Mass Spectrometry Imaging and Integration with Other Imaging Modalities for Greater Molecular Understanding of Biological Tissues. Mol. Imaging Biol. 2018, 20, 888–901. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriegsmann, K.; Longuespee, R.; Hundemer, M.; Zgorzelski, C.; Casadonte, R.; Schwamborn, K.; Weichert, W.; Schirmacher, P.; Harms, A.; Kazdal, D.; et al. Combined Immunohistochemistry after Mass Spectrometry Imaging for Superior Spatial Information. Proteom. Clin. Appl. 2019, 13, e1800035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kriegsmann, M.; Casadonte, R.; Kriegsmann, J.; Dienemann, H.; Schirmacher, P.; Kobarg, J.H.; Schwamborn, K.; Stenzinger, A.; Warth, A.; Weichert, W. Reliable Entity Subtyping in Non-small Cell Lung Cancer by Matrix-assisted Laser Desorption/Ionization Imaging Mass Spectrometry on Formalin-fixed Paraffin-embedded Tissue Specimens. Mol. Cell Proteom. 2016, 15, 3081–3089. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Groseclose, M.R.; Massion, P.P.; Chaurand, P.; Caprioli, R.M. High-throughput proteomic analysis of formalin-fixed paraffin-embedded tissue microarrays using MALDI imaging mass spectrometry. Proteomics 2008, 8, 3715–3724. [Google Scholar] [CrossRef]
- Casadonte, R.; Kriegsmann, M.; Perren, A.; Baretton, G.; Deininger, S.O.; Kriegsmann, K.; Welsch, T.; Pilarsky, C.; Kriegsmann, J. Development of a Class Prediction Model to Discriminate Pancreatic Ductal Adenocarcinoma from Pancreatic Neuroendocrine Tumor by MALDI Mass Spectrometry Imaging. Proteom. Clin. Appl. 2019, 13, e1800046. [Google Scholar] [CrossRef]
- Ly, A.; Longuespee, R.; Casadonte, R.; Wandernoth, P.; Schwamborn, K.; Bollwein, C.; Marsching, C.; Kriegsmann, K.; Hopf, C.; Weichert, W.; et al. Site-to-Site Reproducibility and Spatial Resolution in MALDI-MSI of Peptides from Formalin-Fixed Paraffin-Embedded Samples. Proteom. Clin. Appl. 2019, 13, e1800029. [Google Scholar] [CrossRef] [Green Version]
- Boskamp, T.; Casadonte, R.; Hauberg-Lotte, L.; Deininger, S.; Kriegsmann, J.; Maass, P. Cross-Normalization of MALDI Mass Spectrometry Imaging Data Improves Site-to-Site Reproducibility. Anal. Chem. 2021, 93, 10584–10592. [Google Scholar] [CrossRef]
- Dragovich, T. Pancratic Cancer. Available online: https://emedicine.medscape.com/article/280605-overview (accessed on 30 May 2019).
- Bledsoe, J.R.; Shinagare, S.A.; Deshpande, V. Difficult Diagnostic Problems in Pancreatobiliary Neoplasia. Arch. Pathol. Lab. Med. 2015, 139, 848–857. [Google Scholar] [CrossRef] [Green Version]
- Deshpande, V.; Konstantinidis, I.T.; Castillo, C.F.; Hezel, A.F.; Haigis, K.M.; Ting, D.T.; Bardeesy, N.; Goyal, L.; Zhu, A.X.; Warshaw, A.L.; et al. Intra-pancreatic Distal Bile Duct Carcinoma is Morphologically, Genetically, and Clinically Distinct from Pancreatic Ductal Adenocarcinoma. J. Gastrointest. Surg. 2016, 20, 953–959. [Google Scholar] [CrossRef]
- Brindley, P.J.; Bachini, M.; Ilyas, S.I.; Khan, S.A.; Loukas, A.; Sirica, A.E.; Teh, B.T.; Wongkham, S.; Gores, G.J. Cholangiocarcinoma. Nat. Rev. Dis. Primers 2021, 7, 65. [Google Scholar] [CrossRef]
- Avila-Lopez, P.A.; Guerrero, G.; Nunez-Martinez, H.N.; Peralta-Alvarez, C.A.; Hernandez-Montes, G.; Alvarez-Hilario, L.G.; Herrera-Goepfert, R.; Albores-Saavedra, J.; Villegas-Sepulveda, N.; Cedillo-Barron, L.; et al. H2A.Z overexpression suppresses senescence and chemosensitivity in pancreatic ductal adenocarcinoma. Oncogene 2021, 40, 2065–2080. [Google Scholar] [CrossRef]
- Tian, C.; Clauser, K.R.; Ohlund, D.; Rickelt, S.; Huang, Y.; Gupta, M.; Mani, D.R.; Carr, S.A.; Tuveson, D.A.; Hynes, R.O. Proteomic analyses of ECM during pancreatic ductal adenocarcinoma progression reveal different contributions by tumor and stromal cells. Proc. Natl. Acad. Sci. USA 2019, 116, 19609–19618. [Google Scholar] [CrossRef] [Green Version]
- Tian, C.; Huang, Y.; Clauser, K.R.; Rickelt, S.; Lau, A.N.; Carr, S.A.; Heiden, M.G.V.; Hynes, R.O. Suppression of pancreatic ductal adenocarcinoma growth and metastasis by fibrillar collagens produced selectively by tumor cells. Nat. Commun. 2021, 12, 2328. [Google Scholar] [CrossRef] [PubMed]
- Loch, F.N.; Klein, O.; Beyer, K.; Klauschen, F.; Schineis, C.; Lauscher, J.C.; Margonis, G.A.; Degro, C.E.; Rayya, W.; Kamphues, C. Peptide Signatures for Prognostic Markers of Pancreatic Cancer by MALDI Mass Spectrometry Imaging. Biology 2021, 10, 1033. [Google Scholar] [CrossRef]
- Kalsch, J.; Padden, J.; Bertram, S.; Pott, L.L.; Reis, H.; Westerwick, D.; Schaefer, C.M.; Sowa, J.P.; Mollmann, D.; Fingas, C.; et al. Annexin A10 optimally differentiates between intrahepatic cholangiocarcinoma and hepatic metastases of pancreatic ductal adenocarcinoma: A comparative study of immunohistochemical markers and panels. Virchows Arch. 2017, 470, 537–543. [Google Scholar] [CrossRef] [PubMed]
- Padden, J.; Ahrens, M.; Kalsch, J.; Bertram, S.; Megger, D.A.; Bracht, T.; Eisenacher, M.; Kocabayoglu, P.; Meyer, H.E.; Sipos, B.; et al. Immunohistochemical Markers Distinguishing Cholangiocellular Carcinoma (CCC) from Pancreatic Ductal Adenocarcinoma (PDAC) Discovered by Proteomic Analysis of Microdissected Cells. Mol. Cell Proteom. 2016, 15, 1072–1082. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lok, T.; Chen, L.; Lin, F.; Wang, H.L. Immunohistochemical distinction between intrahepatic cholangiocarcinoma and pancreatic ductal adenocarcinoma. Hum. Pathol. 2014, 45, 394–400. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hooper, J.E.; Morgan, T.K.; Grompe, M.; Sheppard, B.C.; Troxell, M.L.; Corless, C.L.; Streeter, P.R. The novel monoclonal antibody HPC2 and N-cadherin distinguish pancreatic ductal adenocarcinoma from cholangiocarcinoma. Hum. Pathol. 2012, 43, 1583–1589. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hrudka, J.; Prouzova, Z.; Mydlikova, K.; Jedlickova, K.; Holesta, M.; Whitley, A.; Havluj, L. FOXF1 as an Immunohistochemical Marker of Hilar Cholangiocarcinoma or Metastatic Pancreatic Ductal Adenocarcinoma. Single Institution Experience. Pathol. Oncol. Res. 2021, 27, 1609756. [Google Scholar] [CrossRef]
- Somoracz, A.; Tatrai, P.; Horvath, G.; Kiss, A.; Kupcsulik, P.; Kovalszky, I.; Schaff, Z. Agrin immunohistochemistry facilitates the determination of primary versus metastatic origin of liver carcinomas. Hum. Pathol. 2010, 41, 1310–1319. [Google Scholar] [CrossRef]
- Fasanella, K.E.; Krasinskas, A.; Schoedel, K.E.; Sasatomi, E.; Slivka, A.; Whitcomb, D.C.; Sanders, M.; Nodit, L.; Raab, S.; McGrath, K.M.; et al. DNA mutational differences in cytological specimens from pancreatic cancer and cholangiocarcinoma. Pancreatology 2010, 10, 429–433. [Google Scholar] [CrossRef] [PubMed]
- Mosbeh, A.; Halfawy, K.; Abdel-Mageed, W.S.; Sweed, D.; Rahman, M.H.A. Nuclear BAP1 loss is common in intrahepatic cholangiocarcinoma and a subtype of hepatocellular carcinoma but rare in pancreatic ductal adenocarcinoma. Cancer Genet. 2018, 224–225, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Kendall, T.; Verheij, J.; Gaudio, E.; Evert, M.; Guido, M.; Goeppert, B.; Carpino, G. Anatomical, histomorphological and molecular classification of cholangiocarcinoma. Liver Int. 2019, 39 (Suppl. 1), 7–18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Zhang, L.; Bluth, M.H.; Bhalla, A. Molecular Diagnostics in the Neoplasms of the Pancreas, Liver, Gallbladder, and Extrahepatic Biliary Tract: 2018 Update. Clin. Lab. Med. 2018, 38, 367–384. [Google Scholar] [CrossRef] [PubMed]



| Pancreatic Carcinoma (PDAC) (n = 41) | Cholangiocarcinoma (CC) (n = 41) | p Value | |
|---|---|---|---|
| Age in years (mean ± SD) | 66 ± 11.4 | 64 ± 11.9 | 0.49 † |
| Male sex (%) | 73.2 | 51.2 | 0.04 *‡ |
| T stage | |||
| pT1/pT2 (%) | 26.8 | 70.7 | < 0.01 *‡ |
| pT3/pT4 (%) | 73.2 | 29.3 | |
| Nodal involvement | |||
| pN+ (%) | 95.1 | 63.4 | < 0.01 *‡ |
| pN− (%) | 4.9 | 36.6 | |
| Tumor grading | |||
| G1/G2 (%) | 51.2 | 51.2 | 1.00 ‡ |
| G3 (%) | 48.8 | 48.8 | |
| Resection margins | |||
| R0 (%) | 53.7 | 73.2 | 0.07 ‡ |
| R1 (%) | 46.3 | 26.8 |
| Classification Algorithm | Accuracy | FNR * | FPR ** | TNR *** | TPR **** | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Class | CC | PDAC | CC | PDAC | CC | PDAC | CC | PDAC | CC | PDAC |
| Support vector machine | 0.91 | 0.91 | 0.03 | 0.24 | 0.24 | 0.03 | 0.76 | 0.97 | 0.97 | 0.76 |
| Gradient boosting | 0.88 | 0.88 | 0.02 | 0.34 | 0.34 | 0.02 | 0.67 | 0.98 | 0.98 | 0.67 |
| K-nearest neighbors | 0.86 | 0.86 | 0.14 | 0.14 | 0.14 | 0.14 | 0.86 | 0.87 | 0.87 | 0.86 |
| Classification Algorithm | Accuracy | FNR * | FPR ** | TNR *** | TPR **** | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| Class | CC | PDAC | CC | PDAC | CC | PDAC | CC | PDAC | CC | PDAC |
| Support vector machine | 0.89 | 0.89 | 0.16 | 0.07 | 0.07 | 0.16 | 0.93 | 0.84 | 0.84 | 0.93 |
| Gradient boosting | 0.88 | 0.88 | 0.19 | 0.07 | 0.07 | 0.19 | 0.93 | 0.82 | 0.82 | 0.93 |
| K-nearest neighbors | 0.85 | 0.85 | 0.26 | 0.07 | 0.07 | 0.26 | 0.93 | 0.74 | 0.74 | 0.93 |
| Observed m/z | Mr (Expect) | Mr (Calc) | Error (Da) | Peptide Sequence | Protein (UniProtKB Protein Accession Number) | Modifications |
|---|---|---|---|---|---|---|
| 850.4 | 850.5 | 849.5 | 0.03 | R.HLQLAIR.N | Histone H2A (B2R5B3) | |
| 944.5 | 944.6 | 943.5 | 0.10 | R.AGLQFPVGR.I | Histone H2A (B2R5B3) | |
| 1105.5 | 1105.5 | 1104.6 | 0.17 | R.GVQGPPGPAGPR.G | Collagen alpha-1(I) chain (P02452) | Oxidation (p) |
| 2056.0 | 2056.0 | 2056.0 | 0.43 | K.TGPPGPAGQDGRPGPPGPPGAR.G | Collagen alpha-1(I) chain (P02452) | Oxidation (p) |
| 2073.0 | 2073.0 | 2072.0 | 0.24 | K.GSPGADGPAGAPGTPGPQGIAGQR.G | Collagen alpha-1(I) chain(P02452) | |
| 1198.7 | 1198.7 | 1197.7 | 0.22 | R. AVFPSIVGRPR.H | Actin * |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Bollwein, C.; Gonҫalves, J.P.L.; Utpatel, K.; Weichert, W.; Schwamborn, K. MALDI Mass Spectrometry Imaging for the Distinction of Adenocarcinomas of the Pancreas and Biliary Tree. Molecules 2022, 27, 3464. https://doi.org/10.3390/molecules27113464
Bollwein C, Gonҫalves JPL, Utpatel K, Weichert W, Schwamborn K. MALDI Mass Spectrometry Imaging for the Distinction of Adenocarcinomas of the Pancreas and Biliary Tree. Molecules. 2022; 27(11):3464. https://doi.org/10.3390/molecules27113464
Chicago/Turabian StyleBollwein, Christine, Juliana Pereira Lopes Gonҫalves, Kirsten Utpatel, Wilko Weichert, and Kristina Schwamborn. 2022. "MALDI Mass Spectrometry Imaging for the Distinction of Adenocarcinomas of the Pancreas and Biliary Tree" Molecules 27, no. 11: 3464. https://doi.org/10.3390/molecules27113464
APA StyleBollwein, C., Gonҫalves, J. P. L., Utpatel, K., Weichert, W., & Schwamborn, K. (2022). MALDI Mass Spectrometry Imaging for the Distinction of Adenocarcinomas of the Pancreas and Biliary Tree. Molecules, 27(11), 3464. https://doi.org/10.3390/molecules27113464