Molecular Dynamic Study of Mechanism Underlying Nature of Molecular Recognition and the Role of Crosslinker in the Synthesis of Salmeterol-Targeting Molecularly Imprinted Polymer for Analysis of Salmeterol Xinafoate in Biological Fluid
Abstract
:1. Introduction
2. Materials and Methods
2.1. Materials
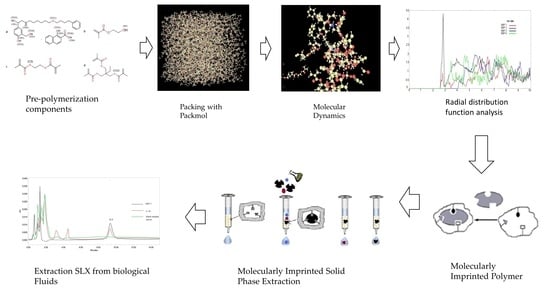
2.2. Molecular Dynamic Simulations
2.3. Synthesis of Molecularly Imprinted Polymer (MIP) of Salmeterol Xinafoate (SLX) and Non-Imprinted Polymer (NIP) Using Precipitation Polymerization
2.4. Evaluation of MIP and NIP Adsorption Capacity
2.5. Optimization of the MISPE Condition
2.6. Application of Molecularly Imprinted Solid Phase Extraction (MISPE) and Non-Imprinted Solid Phase Extraction (NISPE) on Spiked Blood Serum
2.7. Physical Characterization of Sorbent with Fourier Transform Infrared (FTIR), Brunauer-Emmett-Teller (BET), and Scanning Electron Microscope (SEM)
3. Results and Discussion
3.1. Molecular Dynamic Simulations
3.2. Synthesis of the Salmeterol Xinafoate-Imprinted Polymer Using Precipitation Method
3.3. Evaluation of MIP and NIP Adsorption Capacity
3.4. MISPE Optimization
3.5. Effect of Concentration of SLX on % Recovery
3.6. Selectivity Test
3.7. Application of MISPE
3.8. Physical Characterization of Sorbent with Brunauer-Emmett-Teller (BET), Scanning Electron Microscopy (SEM), and Fourier Transform Infrared (FTIR)
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| SLX | Salmeterol xinafoate |
| WADA | World Anti-Doping Agency |
| IOC | International Olympic Committee |
| MIPs | Molecularly imprinted polymers |
| TER | Terbutaline |
| SAL | Salbutamol |
| HEMA | Hydroxyethyl methacrylate |
| EGDMA | Ethylene glycol dimethacrylate |
| TRIM | Trimethylolpropane trimethacrylate |
| BPO | Benzoyl peroxide |
| NIPs | Non-imprinted polymers |
| MISPE | Molecularly imprinted solid phase extraction |
| NISPE | Non-imprinted solid phase extraction |
| HPLC | High-performance liquid chromatography |
| FTIR | Fourier transform infrared |
| SEM | Scanning electron microscope |
| BET | Brunauer-Emmett-Teller |
| T | Template |
| FM | Functional monomer |
| RDF | Radial distribution factor |
| VDW | Van der Waals |
References
- Kondawar, M.S.; Shah, R.R.; Waghmare, J.J.; Malusare, M.K.; Shah, N.D. UV spectrophotometric method for simultaneous estimation of Salmeterol xinafoate and Fluticasone propionate in Bulk and Dosage form. Int. J. PharmTech Res. 2011, 3, 1801–1806. [Google Scholar]
- Pluim, B.M.; De Hon, O.; Staal, B.; Limpens, J.; Kuipers, H.; Overbeek, S.E.; Zwinderman, A.H.; Scholten, R.J.P.M. β2-Agonists and Physical Performance. Sports Med. 2011, 41, 39–57. [Google Scholar] [CrossRef] [PubMed]
- Deventer, K.; Pozo, O.; Delbeke, F.T.; Van Eenoo, P. Quantitative Detection of Inhaled Salmeterol in Human Urine and Relevance to Doping Control Analysis. Ther. Drug Monit. 2011, 33, 627–631. [Google Scholar] [CrossRef] [PubMed]
- Fragkaki, A.; Georgakopoulos, C.; Sterk, S.; Nielen, M. Sports doping: Emerging designer and therapeutic β2-agonists. Clin. Chim. Acta 2013, 425, 242–258. [Google Scholar] [CrossRef] [PubMed]
- Carlsen, K.-H.; Hem, E.; Stensrud, T.; Held, T.; Herland, K.; Mowinckel, P. Can asthma treatment in sports be doping? The effect of the rapid onset, long-acting inhaledβ2 -agonist formoterol upon endurance performance in healthy well-trained athletes. Respir. Med. 2001, 95, 571–576. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wen, Y.; Chen, L.; Li, J.; Liu, D.; Chen, L. Recent advances in solid-phase sorbents for sample preparation prior to chromatographic analysis. TrAC Trends Anal. Chem. 2014, 59, 26–41. [Google Scholar] [CrossRef]
- Suryana, S.; Mutakin, M.; Rosandi, Y.; Hasanah, A.N. Rational design of salmeterol xinafoate imprinted polymer through computational method: Functional monomer and crosslinker selection. Polym. Adv. Technol. 2021, 33, 221–234. [Google Scholar] [CrossRef]
- Mathew, D.; Thomas, B.; Devaky, K.S. Design, synthesis and characterization of enzyme-analogue-built polymer catalysts as artificial hydrolases. Artif. Cells Nanomed. Biotechnol. 2019, 47, 1149–1172. [Google Scholar] [CrossRef] [Green Version]
- Marć, M.; Wieczorek, P.P. Introduction to MIP synthesis, characteristics and analytical application. In Comprehensive Analytical Chemistry; Elsevier: Amsterdam, The Netherlands, 2019; pp. 1–15. [Google Scholar] [CrossRef]
- Feizy, J.; Nezhadali, A.; Es’Haghi, Z.; Beheshti, H.R. HPLC Determination of Hexythiazox in Food Samples by MISPE Extraction. Chromatographia 2017, 80, 437–446. [Google Scholar] [CrossRef]
- Zeng, H.; Wang, Y.; Liu, X.; Kong, J.; Nie, C. Preparation of molecular imprinted polymers using bi-functional monomer and bi-crosslinker for solid-phase extraction of rutin. Talanta 2012, 93, 172–181. [Google Scholar] [CrossRef]
- Wang, L.; Fu, W.; Shen, Y.; Tan, H.; Xu, H. Molecularly Imprinted Polymers for Selective Extraction of Oblongifolin C from Garcinia yunnanensis Hu. Molecules 2017, 22, 508. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Balamurugan, K.; Gokulakrishnan, K.; Prakasam, T. Preparation and evaluation of molecularly imprinted polymer liquid chromatography column for the separation of Cathine enantiomers. Saudi Pharm. J. 2011, 20, 53–61. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hasanah, A.N.; Maelaningsih, F.S.; Apriliandi, F.; Sabarudin, A. Synthesis and Characterisation of a Monolithic Imprinted Column Using a Methacrylic Acid Monomer with Porogen Propanol for Atenolol Analysis. J. Anal. Methods Chem. 2020, 2020, 3027618. [Google Scholar] [CrossRef] [PubMed]
- Becskereki, G.; Horvai, G.; Tóth, B. The Selectivity of Molecularly Imprinted Polymers. Polymers 2021, 13, 1781. [Google Scholar] [CrossRef] [PubMed]
- Pei, H.Y.; Shen, G.J.; Du, Y. The Application of Molecularly Imprinted Polymers to Solid-Phase Extraction. Adv. Mater. Res. 2014, 893, 283–286. [Google Scholar] [CrossRef]
- Nezhadali, A.; Rouki, Z.; Nezhadali, M. Electrochemical preparation of a molecularly imprinted polypyrrole modified pencil graphite electrode for the determination of phenothiazine in model and real biological samples. Talanta 2015, 144, 456–465. [Google Scholar] [CrossRef]
- González-Vila, A.; Debliquy, M.; Lahem, D.; Zhang, C.; Mégret, P.; Caucheteur, C. Molecularly imprinted electropolymerization on a metal-coated optical fiber for gas sensing applications. Sens. Actuators B Chem. 2017, 244, 1145–1151. [Google Scholar] [CrossRef]
- Olsson, G.D.; Karlsson, B.C.G.; Shoravi, S.; Wiklander, J.G.; Nicholls, I.A. Mechanisms underlying molecularly imprinted polymer molecular memory and the role of crosslinker: Resolving debate on the nature of template recognition in phenylalanine anilide imprinted polymers. J. Mol. Recognit. 2012, 25, 69–73. [Google Scholar] [CrossRef]
- Fernandes, L.S.; Homem-De-Mello, P.; de Lima, E.C.; Honorio, K.M. Rational design of molecularly imprinted polymers for recognition of cannabinoids: A structure–property relationship study. Eur. Polym. J. 2015, 71, 364–371. [Google Scholar] [CrossRef]
- Zhang, W.; She, X.; Wang, L.; Fan, H.; Zhou, Q.; Huang, X.; Tang, J.Z. Preparation, Characterization and Application of a Molecularly Imprinted Polymer for Selective Recognition of Sulpiride. Materials 2017, 10, 475. [Google Scholar] [CrossRef] [Green Version]
- Muhammad, T.; Nur, Z.; Piletska, E.V.; Yimit, O.; Piletsky, S.A. Rational design of molecularly imprinted polymer: The choice of cross-linker. Analyst 2012, 137, 2623–2628. [Google Scholar] [CrossRef] [PubMed]
- Golker, K.; Karlsson, B.C.G.; Rosengren, A.M.; Nicholls, I.A. A Functional Monomer Is Not Enough: Principal Component Analysis of the Influence of Template Complexation in Pre-Polymerization Mixtures on Imprinted Polymer Recognition and Morphology. Int. J. Mol. Sci. 2014, 15, 20572–20584. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Iturralde, I.; Paulis, M.; Leiza, J. The effect of the crosslinking agent on the performance of propranolol imprinted polymers. Eur. Polym. J. 2014, 53, 282–291. [Google Scholar] [CrossRef]
- Qiu, C.; Yang, W.; Zhou, Z.; Yan, Y.; Xu, W. Rational design and preparation of dibenzothiophene-targeting molecularly imprinted polymers with molecular dynamics approaches and surface-initiated activators regenerated by electron-transfer atom-transfer radical polymerization. J. Appl. Polym. Sci. 2015, 132, 42629. [Google Scholar] [CrossRef]
- Mohebali, A.; Abdouss, M.; Mazinani, S.; Zahedi, P. Synthesis and characterization of poly(methacrylic acid)-based molecularly imprinted polymer nanoparticles for controlled release of trinitroglycerin. Polym. Adv. Technol. 2016, 27, 1164–1171. [Google Scholar] [CrossRef]
- Ratautaite, V.; Maruška, A.; Erickson, M.; Kornyšova, O. Effect of polymerization conditions on morphology and chromatographic characteristics of polyacrylamide-based beds (monoliths) for capillary electrochromatography and capillary liquid chromatography. J. Sep. Sci. 2009, 32, 2582–2591. [Google Scholar] [CrossRef]
- Kornyšova, O.; Jarmalavičienė, R.; Maruška, A. A simplified synthesis of polymeric nonparticulate stationary phases with macrocyclic antibiotic as chiral selector for capillary electrochromatography. Electrophoresis 2004, 25, 2825–2829. [Google Scholar] [CrossRef]
- Zhang, M.; Huang, J.; Yu, P.; Chen, X. Preparation and characteristics of protein molecularly imprinted membranes on the surface of multiwalled carbon nanotubes. Talanta 2010, 81, 162–166. [Google Scholar] [CrossRef]
- Shinde, S.; Incel, A.; Mansour, M.; Olsson, G.D.; Nicholls, I.A.; Esen, C.; Urraca, J.; Sellergren, B. Urea-Based Imprinted Polymer Hosts with Switchable Anion Preference. J. Am. Chem. Soc. 2020, 142, 11404–11416. [Google Scholar] [CrossRef]
- Lusina, A.; Cegłowski, M. Molecularly Imprinted Polymers as State-of-the-Art Drug Carriers in Hydrogel Transdermal Drug Delivery Applications. Polymers 2022, 14, 640. [Google Scholar] [CrossRef]
- LeJeune, J.; Spivak, D.A. Chiral effects of alkyl-substituted derivatives of N,O-bismethacryloyl ethanolamine on the performance of one monomer molecularly imprinted polymers (OMNiMIPs). Anal. Bioanal. Chem. 2007, 389, 433–440. [Google Scholar] [CrossRef]
- Zhang, J.; Wang, L.-L.; Ma, J.-Q.; Wang, Y.-L. Preparation of Ofloxacin Poly(Glycidyl Methacrylate-Co-Ethylenedimethacrylate) (PGMA/EDMA) Molecularly Imprinted Microspheres and Their Application to the Analysis of Quinolones in Milk. Food Anal. Methods 2013, 7, 721–729. [Google Scholar] [CrossRef]
- Bunina, Z.; Bryleva, K.; Belikov, K. Synthesis and Adsorption Properties of Gadolinium-Imprinted Divinylbenzene-Based Copolymers. ACS Omega 2021, 6, 3336–3344. [Google Scholar] [CrossRef] [PubMed]
- Ghani, N.T.A.; Abdulla, H.; Rizk, M.S.; Dena, A.S.A.; El Nashar, R.M. Molecularly imprinted polymer/reduced graphene oxide–based carbon–paste sensor for highly sensitive determination of the anti–HCV drug daclatasvir dihydrochloride. Sens. Actuators B: Chem. 2018, 283, 6–17. [Google Scholar] [CrossRef]
- Chopra, I.; Rahangdale, D.; Kumar, A. Computational modeling for rational designing of imprinted polymers for herbicides: A review. Indian J. Agric. Sci. 2019, 89, 1063–1070. [Google Scholar]
- Cleland, D.; Olsson, G.D.; Karlsson, B.C.G.; Nicholls, I.A.; McCluskey, A. Molecular dynamics approaches to the design and synthesis of PCB targeting molecularly imprinted polymers: Interference to monomer–template interactions in imprinting of 1,2,3-trichlorobenzene. Org. Biomol. Chem. 2013, 12, 844–853. [Google Scholar] [CrossRef]
- Pardeshi, S.; Singh, S.K. Precipitation polymerization: A versatile tool for preparing molecularly imprinted polymer beads for chromatography applications. RSC Adv. 2016, 6, 23525–23536. [Google Scholar] [CrossRef]
- Hasanah, A.N.; Rahayu, D.; Pratiwi, R.; Rostinawati, T.; Megantara, S.; Saputri, F.A.; Puspanegara, K.H. Extraction of atenolol from spiked blood serum using a molecularly imprinted polymer sorbent obtained by precipitation polymerization. Heliyon 2019, 5, e01533. [Google Scholar] [CrossRef] [Green Version]
- Vanommeslaeghe, K.; Hatcher, E.; Acharya, C.; Kundu, S.; Zhong, S.; Shim, J.; Darian, E.; Guvench, O.; Lopes, P.; Vorobyov, I.; et al. CHARMM General Force Field: A Force Field for Drug-Like Molecules Compatible with the CHARMM All-Atom Additive Biological Force Fields. J. Comput. Chem. 2010, 31, 671–690. [Google Scholar] [CrossRef] [Green Version]
- Crozier, P.; Plimpton, S.; Thompson, A.; Brown, M. A brief introduction to MD. In A Brief Survey of the LAMMPS MD Code: Intro, Case Studies, and Future Development; Sandia National Laboratories: Albuquerque, NM, USA, 2010. [Google Scholar]
- Senftle, T.P.; Hong, S.; Islam, M.; Kylasa, S.B.; Zheng, Y.; Shin, Y.K.; Junkermeier, C.; Engel-Herbert, R.; Janik, M.J.; Aktulga, H.M.; et al. The ReaxFF Reactive Force-Field: Development, Applications and Future Directions. Comput. Mater. 2016, 2, 15011. [Google Scholar] [CrossRef]
- Pratama, K.F.; Manik, M.E.R.; Rahayu, D.; Hasanah, A.N. Effect of the Molecularly Imprinted Polymer Component Ratio on Analytical Performance. Chem. Pharm. Bull. 2020, 68, 1013–1024. [Google Scholar] [CrossRef] [PubMed]
- Grabowski, S.J. What Is the Covalency of Hydrogen Bonding? Chem. Rev. 2011, 111, 2597–2625. [Google Scholar] [CrossRef] [PubMed]
- Ali, H.R.H.; Edwards, H.G.; Hargreaves, M.D.; Munshi, T.; Scowen, I.J.; Telford, R.J. Vibrational spectroscopic characterisation of salmeterol xinafoate polymorphs and a preliminary investigation of their transformation using simultaneous in situ portable Raman spectroscopy and differential scanning calorimetry. Anal. Chim. Acta 2008, 620, 103–112. [Google Scholar] [CrossRef] [PubMed]
- Shoravi, S.; Olsson, G.D.; Karlsson, B.C.G.; Nicholls, I.A. On the Influence of Crosslinker on Template Complexation in Molecularly Imprinted Polymers: A Computational Study of Prepolymerization Mixture Events with Correlations to Template-Polymer Recognition Behavior and NMR Spectroscopic Studies. Int. J. Mol. Sci. 2014, 15, 10622–10634. [Google Scholar] [CrossRef] [Green Version]
- Klein, R.A. Modified van der Waals atomic radii for hydrogen bonding based on electron density topology. Chem. Phys. Lett. 2006, 425, 128–133. [Google Scholar] [CrossRef]
- Zhang, I.Y.; Wu, J.; Xu, X. Extending the reliability and applicability of B3LYP. Chem. Commun. 2010, 46, 3057–3070. [Google Scholar] [CrossRef] [Green Version]
- Fisher, T.R.; Zhou, G.; Shi, Y.; Huang, L. How does hydrogen bond network analysis reveal the golden ratio of water–glycerol mixtures? Phys. Chem. Chem. Phys. 2020, 22, 2887–2907. [Google Scholar] [CrossRef]
- Pappalardo, A.; E Amato, M.; Ballistreri, F.P.; Fragola, V.L.P.; A Tomaselli, G.; Toscano, R.M.; Sfrazzetto, G.T. Binding of reactive organophosphate by oximes via hydrogen bond. J. Chem. Sci. 2013, 125, 869–873. [Google Scholar] [CrossRef]
- Estarellas, C.; Escudero, D.; Frontera, A.; Quiñonero, D.; Deyà, P.M. Theoretical ab initio study of the interplay between hydrogen bonding, cation–π and π–π interactions. Theor. Chim. Acta 2009, 122, 325–332. [Google Scholar] [CrossRef]
- Wang, J.; Cormack, P.A.G.; Sherrington, D.C.; Khoshdel, E. Monodisperse, Molecularly Imprinted Polymer Microspheres Prepared by Precipitation Polymerization for Affinity Separation Applications. Angew. Chem. 2003, 115, 5494–5496. [Google Scholar] [CrossRef]
- Adumitrăchioaie, A. Electrochemical Methods Based on Molecularly Imprinted Polymers for Drug Detection. A Review. Int. J. Electrochem. Sci. 2018, 2556–2576. [Google Scholar] [CrossRef]
- Yang, W.; Ma, P.; Fan, T.; Zhou, Z.; Liu, H.; Xu, W. Optimal design of an imprinted preassembled system by quantum chemical calculations and preparation of a surface-imprinted material for the selective removal of quinoline. J. Appl. Polym. Sci. 2014, 132, 41730. [Google Scholar] [CrossRef]
- Wang, R.; Pan, J.; Qin, M.; Guo, T. Molecularly imprinted nanocapsule mimicking phosphotriesterase for the catalytic hydrolysis of organophosphorus pesticides. Eur. Polym. J. 2018, 110, 1–8. [Google Scholar] [CrossRef]
- Li, H.; Chen, J.; Tan, L.; Wang, J. Solid-phase extraction using a molecularly imprinted polymer for the selective purification and preconcentration of norfloxacin from seawater. Anal. Lett. 2019, 52, 2896–2913. [Google Scholar] [CrossRef]
- Maranata, G.J.; Surya, N.O.; Hasanah, A.N. Optimising factors affecting solid phase extraction performances of molecular imprinted polymer as recent sample preparation technique. Heliyon 2021, 7, e05934. [Google Scholar] [CrossRef] [PubMed]
- Koohpaei, A.R.; Shahtaheri, S.J.; Ganjali, M.R.; Forushani, A.R.; Golbabaei, F. Molecular imprinted solid phase extraction for determination of atrazine in environmental samples. Iran. J. Environ. Health Sci. Eng. 2008, 5, 283–296. [Google Scholar]
- Čápka, V.; Carter, S.J. Minimizing matrix effects in the development of a method for the determination of salmeterol in human plasma by LC/MS/MS at low pg/mL concentration levels. J. Chromatogr. B 2007, 856, 285–293. [Google Scholar] [CrossRef]
- Colthup, P.V.; Young, G.C.; Felgate, C.C. Determination of salmeterol in rat and dog plasma by high-performance liquid chromatography with fluorescence detection. J Pharm. Sci. 1993, 82, 323–325. [Google Scholar] [CrossRef]
- Garg, M.; Naidu, R.; Birhade, A.; Iyer, K.; Jadhav, R.; Rebello, J.; Morde, N.; Mangale, M.; Brashier, B. Bioequivalence of Two Formulations of Salmeterol Xinafoate/Fluticasone Propionate HFA pMDI in Healthy Volunteers. J. Bioequiv. Bioavailab. 2017, 9, 536–546. [Google Scholar] [CrossRef]
- Paczkowska, E.; Smukowska, D.; Tratkiewicz, E.; Białasiewicz, P. HPLC method for simultaneous determination of salmeterol xinafoate and fluticasone propionate for the quality control of dry powder inhalation products. Acta Chromatogr. 2015, 27, 309–320. [Google Scholar] [CrossRef] [Green Version]
- Hatami, M.; Karimnia, E.; Farhadi, K. Determination of salmeterol in dried blood spot using an ionic liquid based dispersive liquid–liquid microextraction coupled with HPLC. J. Pharm. Biomed. Anal. 2013, 85, 283–287. [Google Scholar] [CrossRef] [PubMed]
- Silvestro, L.; Savu, S.R.; Savu, S.N.; Tudoroniu, A.; Tarcomnicu, I. Development of a sensitive method for simultaneous determination of fluticasone propionate and salmeterol in plasma samples by liquid chromatography-tandem mass spectrometry. Biomed. Chromatogr. 2011, 26, 627–635. [Google Scholar] [CrossRef] [PubMed]
- Farrington, K.; Magner, E.; Regan, F. Predicting the performance of molecularly imprinted polymers: Selective extraction of caffeine by molecularly imprinted solid phase extraction. Anal. Chim. Acta 2006, 566, 60–68. [Google Scholar] [CrossRef]
- Marć, M.; Wieczorek, P.P. The preparation and evaluation of core-shell magnetic dummy-template molecularly imprinted polymers for preliminary recognition of the low-mass polybrominated diphenyl ethers from aqueous solutions. Sci. Total Environ. 2020, 724, 138151. [Google Scholar] [CrossRef]
- Lay, S.; Yu, H.-N.; Hu, B.-X.; Shen, S.-R. Molecularly imprinted polymers as the extracted sorbents of clenbuterol ahead of liquid chromatographic determination. J. Zhejiang Univ. Sci. B 2016, 17, 465–475. [Google Scholar] [CrossRef] [Green Version]
- Mehdipour, M.; Ansari, M.; Pournamdari, M.; Zeidabadinejad, L.; Kazemipour, M. Selective extraction of organophosphorous pesticides in plasma by magnetic molecularly imprinted polymers with the aid of computational design. Anal. Methods 2018, 10, 4136–4142. [Google Scholar] [CrossRef]











| Polymer | TO1-MH | TO2-MH | TO3-MH | TO4-MH | TO5-MH | TO6-MH | TO7-MH | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| r (Å) | g (r) | r (Å) | g (r) | r (Å) | g (r) | r (Å) | g (r) | r (Å) | g (r) | r (Å) | g (r) | r (Å) | g (r) | |
| MIP1 | 2.7386 | 0.887 | 3.040 | 1.957 | 2.889 | 0.925 | 3.1909 | 2.737 | 2.7386 | 2.324 | - | - | 2.6884 | 0.783 |
| MIP2 | - | - | - | - | 3.1407 | 0.083 | 2.7889 | 0.7708 | 3.1909 | 3.438 | - | - | 3.1909 | 0.316 |
| MIP3 | 2.7885 | 4.328 | 2.939 | 6.125 | 2.7386 | 6.658 | 3.1407 | 4.863 | 2.7889 | 4,386 | 3.2 | 0.175 | 2.7386 | 3.895 |
| MIP4 | 3.1407 | 0.486 | 2.638 | 1.951 | 2.9899 | 4.449 | 3.0904 | 2.9624 | 2.839 | 2.295 | - | - | 2.6381 | 0.675 |
| Polymer | Langmuir | Freundlich | ||||
|---|---|---|---|---|---|---|
| R2 | KL (L/mg) | Qm (mg/g) | R2 | m | a (mg/g) | |
| MIP 1 | 0.9954 | 0.1409 | 2.8216 | 0.9998 | 0.5691 | 0.4640 |
| NIP 1 | 0.8289 | 0.0317 | 2.4850 | 0.9918 | 0.8025 | 0.0944 |
| MIP 2 | 0.9882 | 0.0770 | 4.6232 | 0.9994 | 0.7555 | 0.3801 |
| NIP 2 | 0.8859 | 0.0226 | 3.0450 | 0.9948 | 0.8593 | 0.0781 |
| MIP 3 | 0.9997 | 0.2721 | 3.6751 | 0.9997 | 0.2721 | 0.8908 |
| NIP 3 | 0.9958 | 0.9573 | 1.0446 | 0.9958 | 0.9573 | 0.0865 |
| MIP 4 | 0.999 | 0.346 | 2.8901 | 0.999 | 0.346 | 0.7025 |
| NIP 4 | 0.991 | 0.9714 | 1.0294 | 0.991 | 0.9714 | 0.0902 |
| Target | Preparation Sample | Instrument | Repeatability | % Recovery | Limit of Quantification | Reference |
|---|---|---|---|---|---|---|
| Salmeterol | SPE | HPLC | 3.7–16.3% | 74–84% | 2.0 ng/ml | [60] |
| Salmeterol xinafoate | SPE | LC-MS/MS | 1.86–6.12% | 98.31–100.00% | 2.0 pg/mL | [61] |
| Salmeterol xinafoate | LLE | HPLC | >2.2% | 98.2–102.7% | 0.025 μg/mL−1 | [62] |
| Salmeterol xinafoate | LLME | HPLC | 6.0–8.5% | >90.0% | 0.30 ng/ mL | [63] |
| Salmeterol xinafoate | LLE | LC-MS | 8.8–13.7% | 103.6% | 2.5 pg/mL | [64] |
| Salmeterol xinafoate | MISPE | HPLC | 1.7–6.8% | 98.7% | 6.3 ng/ml | Our result |
| Polymer | Surface Area (m2/g) | |
|---|---|---|
| Precipitation Method | Bulk Method [7] | |
| MIP1 | 254.419 | 42.297 |
| NIP1 | 23.051 | 18.367 |
| MIP2 | 211.486 | 40.674 |
| NIP2 | 35.393 | 6.033 |
| MIP3 | 291.706 | 221.757 |
| NIP3 | 40.216 | 61.381 |
| MIP4 | 230.160 | 141.370 |
| NIP4 | 32.882 | 37.131 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Suryana, S.; Mutakin, M.; Rosandi, Y.; Hasanah, A.N. Molecular Dynamic Study of Mechanism Underlying Nature of Molecular Recognition and the Role of Crosslinker in the Synthesis of Salmeterol-Targeting Molecularly Imprinted Polymer for Analysis of Salmeterol Xinafoate in Biological Fluid. Molecules 2022, 27, 3619. https://doi.org/10.3390/molecules27113619
Suryana S, Mutakin M, Rosandi Y, Hasanah AN. Molecular Dynamic Study of Mechanism Underlying Nature of Molecular Recognition and the Role of Crosslinker in the Synthesis of Salmeterol-Targeting Molecularly Imprinted Polymer for Analysis of Salmeterol Xinafoate in Biological Fluid. Molecules. 2022; 27(11):3619. https://doi.org/10.3390/molecules27113619
Chicago/Turabian StyleSuryana, Shendi, Mutakin Mutakin, Yudi Rosandi, and Aliya Nur Hasanah. 2022. "Molecular Dynamic Study of Mechanism Underlying Nature of Molecular Recognition and the Role of Crosslinker in the Synthesis of Salmeterol-Targeting Molecularly Imprinted Polymer for Analysis of Salmeterol Xinafoate in Biological Fluid" Molecules 27, no. 11: 3619. https://doi.org/10.3390/molecules27113619
APA StyleSuryana, S., Mutakin, M., Rosandi, Y., & Hasanah, A. N. (2022). Molecular Dynamic Study of Mechanism Underlying Nature of Molecular Recognition and the Role of Crosslinker in the Synthesis of Salmeterol-Targeting Molecularly Imprinted Polymer for Analysis of Salmeterol Xinafoate in Biological Fluid. Molecules, 27(11), 3619. https://doi.org/10.3390/molecules27113619