Sesquiterpenes from Artemisia annua and Their Cytotoxic Activities
Abstract
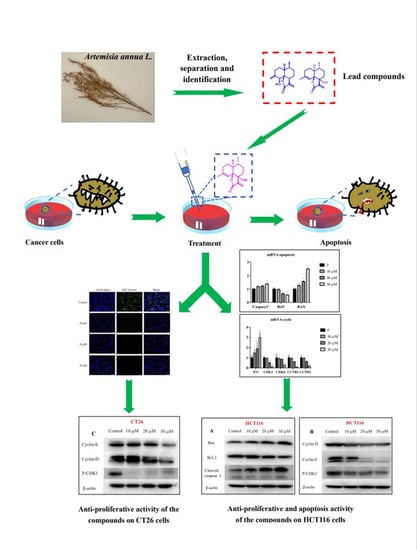
:1. Introduction
2. Results and Discussion
3. Materials and Methods
3.1. General Experimental Procedures
3.2. Plant Material
3.3. Extraction and Isolation
3.4. Quantum Chemical NMR and ECD Calculations of Compound 1–2
3.5. Cell Culture
3.6. Determination of Cell Viability by CCK-8 Assay
3.7. Quantitative Real-Time Polymerase Chain Reaction (q-PCR) Analysis
3.8. EdU Staining Assay
3.9. Western Blotting Assay
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Wang, H.; Li, H.; Zeng, F.L.; Xie, C.X. Spatial distribution of Artemisia annua and potential climate-friendly areas in the world. Chin. Mater. Med. 2015, 38, 460–466. [Google Scholar]
- Shang, Z.J. “Hao, Qinghao, Baihao” in “Fifty-two Diseases Prescriptions”. Chin. Mater. Med. 1988, 42. [Google Scholar] [CrossRef]
- Zhang, X.B.; Zhao, Y.P.; Huang, X.W. Review on study of Dao-di herbs Artemisia annua herba. Chin. J. Chin. Mater. Med. 2016, 41, 2015–2018. [Google Scholar]
- Tu, Y. Artemisinin-A Gift from Traditional Chinese Medicine to the World (Nobel Lecture). Angew. Chem. Int. Ed. Engl. 2016, 55, 10210–10226. [Google Scholar] [CrossRef] [PubMed]
- Feng, X.; Cao, S.; Qiu, F.; Zhang, B. Traditional application and modern pharmacological research of Artemisia annua L. Pharmacol. Ther. 2020, 216, 107650. [Google Scholar] [CrossRef]
- Zhigzhitzhapova, S.V.; Dylenova, E.P.; Gulyaev, S.M.; Randalova, T.E.; Taraskin, V.V.; Tykheev, Z.A.; Radnaeva, L.D. Composition and antioxidant activity of the essential oil of Artemisia annua L. Nat. Prod. Res. 2020, 34, 2668–2671. [Google Scholar] [CrossRef]
- Liu, X.; Cao, J.; Huang, G.; Zhao, Q.; Shen, J. Biological Activities of Artemisinin Derivatives Beyond Malaria. Curr. Top. Med. Chem. 2019, 19, 205–222. [Google Scholar] [CrossRef]
- Efferth, T. Willmar Schwabe Award 2006: Antiplasmodial and antitumor activity of artemisinin--from bench to bedside. Planta. Med. 2007, 73, 299–309. [Google Scholar] [CrossRef] [Green Version]
- Septembre-Malaterre, A.; Lalarizo-Rakoto, M.; Marodon, C.; Bedoui, Y.; Nakab, J.; Simon, E.; Hoarau, L.; Savriama, S.; Strasberg, D.; Guiraud, P.; et al. Artemisia annua, a Traditional Plant Brought to Light. Int. J. Mol. Sci. 2020, 21, 4986. [Google Scholar] [CrossRef]
- Tayarani-Najaran, Z.; Akaberi, M.; Hassanzadeh, B.; Shirazi, N.; Asili, J.; Al-Najjar, H.; Sahebkar, A.; Emami, S.A. Analysis of the Essential Oils of Five Artemisia Species and Evaluation of their Cytotoxic and Proapoptotic Effects. Mini. Rev. Med. Chem. 2019, 19, 902–912. [Google Scholar] [CrossRef]
- Mancuso, R.I.; Foglio, M.A.; Olalla-Saad, S.T. Artemisinin-type drugs for the treatment of hematological malignancies. Cancer. Chemother. Pharmacol. 2021, 1, 1–22. [Google Scholar] [CrossRef]
- Efferth, T. From ancient herb to modern drug: Artemisia annua and artemisinin for cancer therapy. Semin. Cancer. Biol. 2017, 46, 65–83. [Google Scholar] [CrossRef]
- Gao, F.; Sun, Z.; Kong, F.; Xiao, J. Artemisinin-derived hybrids and their anticancer activity. Eur. J. Med. Chem. 2020, 188, 112044. [Google Scholar] [CrossRef]
- Woerdenbag, H.J.; Moskal, T.A.; Pras, N.; Malingré, T.M.; El-Feraly, F.S.; Kampinga, H.H.; Konings, A.W. Cytotoxicity of artemisinin-related endoperoxides to Ehrlich ascites tumor cells. J. Nat. Prod. 1993, 56, 849–856. [Google Scholar] [CrossRef]
- Sun, Q.; Teong, B.; Chen, I.F.; Chang, S.J.; Gao, J.; Kuo, S.M. Enhanced apoptotic effects of dihydroartemisinin-aggregated gelatin and hyaluronan nanoparticles on human lung cancer cells. J. Biomed. Mater. Res. B Appl. Biomater. 2014, 102, 455–462. [Google Scholar] [CrossRef]
- Jamalzadeh, L.; Ghafoori, H.; Aghamaali, M.; Sariri, R. Induction of Apoptosis in Human Breast Cancer MCF-7 Cells by a Semi-Synthetic Derivative of Artemisinin: A Caspase-Related Mechanism. Iran. J. Biotechnol. 2017, 15, 157–165. [Google Scholar] [CrossRef] [Green Version]
- Greenshields, A.L.; Shepherd, T.G.; Hoskin, D.W. Contribution of reactive oxygen species to ovarian cancer cell growth arrest and killing by the anti-malarial drug artesunate. Mol. Carcinog. 2017, 56, 75–93. [Google Scholar] [CrossRef]
- Tran, K.Q.; Tin, A.S.; Firestone, G.L. Artemisinin triggers a G1 cell cycle arrest of human Ishikawa endometrial cancer cells and inhibits cyclin-dependent kinase-4 promoter activity and expression by disrupting nuclear factor-κB transcriptional signaling. Anticancer Drugs 2014, 25, 270–281. [Google Scholar] [CrossRef] [Green Version]
- Zhang, S.; Shi, L.; Ma, H.; Li, H.; Li, Y.; Lu, Y.; Wang, Q.; Li, W. Dihydroartemisinin induces apoptosis in human gastric cancer cell line BGC-823 through activation of JNK1/2 and p38 MAPK signaling pathways. J. Recept. Signal Transduct. Res. 2016, 37, 174–180. [Google Scholar] [CrossRef]
- Takenaka, Y.; Seki, S.; Nishi, T.; Tanahashi, T. Two new sesquiterpenes from Artemisia annua L. J. Nat. Med. 2020, 74, 811–818. [Google Scholar] [CrossRef]
- Li, H.B.; Yu, Y.; Wang, Z.Z.; Yang, J.; Xiao, W.; Yao, X.S. Two new sesquiterpenoids from Artemisia annua. Magn. Reson. Chem. 2015, 53, 244–247. [Google Scholar] [CrossRef] [PubMed]
- Van-Herpen, T.W.; Cankar, K.; Nogueira, M.; Bosch, D.; Bouwmeester, H.J.; Beekwilder, J. Nicotiana benthamiana as a production platform for artemisinin precursors. PLoS ONE 2010, 5, e14222. [Google Scholar] [CrossRef] [PubMed]
- Elmarakby, S.A.; El-Feraly, F.S.; Elsohly, H.N.; Croom, E.M.; Hufford, C.D. Microbial transformation studies on arteannuin B. J. Nat. Prod. 1987, 50, 903–909. [Google Scholar] [CrossRef] [PubMed]
- Lee, I.S.; ElSohly, H.N.; Croom, E.M.; Hufford, C.D. Microbial metabolism studies of the antimalarial sesquiterpene artemisinin. J. Nat. Prod. 1989, 52, 337–341. [Google Scholar] [CrossRef]
- Sy, L.K.; Brown, G.D. Synthesis of 6,7-dehydroartemisinic acid. J. Chem. Soc. 2001, 19, 2421–2429. [Google Scholar] [CrossRef]
- Qin, D.P.; Li, H.B.; Pang, Q.Q.; Huang, Y.X.; Pan, D.B.; Su, Z.Z.; Yao, X.J.; Yao, X.S.; Xiao, W.; Yu, Y. Structurally diverse sesquiterpenoids from the aerial parts of Artemisia annua (Qinghao) and their striking systemically anti-inflammatory activities. Bioorg. Chem. 2020, 103, 104221. [Google Scholar] [CrossRef]
- Heymann, H.; Tezuka, Y.; Kikuchi, T. Constituents of Sindora sumatrana MIQ. I. Isolation and NMR Spectral Analysis of Sesquiterpenes from the Dried Pods. Chem. Pharm. Bull. 1994, 42, 138–146. [Google Scholar] [CrossRef] [Green Version]
- Xu, Y.; Zhang, H.W.; Yu, C.Y.; Lu, Y.; Chang, Y.; Zou, Z.M. Norcyperone, a novel skeleton norsesquiterpene from Cyperus rotundus L. Molecules 2008, 13, 2474–2481. [Google Scholar] [CrossRef]
- Pang, X.Y.; Li, Y.X.; Gong, Y.; Yan, Y.; Li, H.F.; Zhu, Y. Sesquiterpenes from the whole plants of Parasenecio roborowskii. Fitoterapia 2017, 116, 24–33. [Google Scholar] [CrossRef]
- Triana, J.; López, M.; Pérez, F.J.; González-Platas, J.; Quintana, J.; Estévez, F.; León, F.; Bermejo, J. Sesquiterpenoids from Pulicaria canariensis and their cytotoxic activities. J. Nat. Prod. 2005, 68, 523–531. [Google Scholar] [CrossRef]
- Misra, L.N.; Ahmad, A.; Thakur, R.S. Bisnor-cadinanes from Artemisia annua and definitive 13C NMR assignments of β-arteether. Phytochemistry 1993, 33, 1461–1464. [Google Scholar] [CrossRef]
- Sy, L.K.; Brown, G.D. Deoxyarteannuin B, dihydro-deoxyarteannuin B and trans-5-hydroxy-2-isopropenyl-5-methylhex-3-en-1-ol from Artemisia anuua. Phytochemistry 2001, 58, 1159–1166. [Google Scholar] [CrossRef]
- Zhu, W.M.; Zhao, Q.; Li, S.L.; Hao, X.J. Sesquiterpenoids from Hedychium yunnanense and Porana discifera, and the structural revision of two sesquiterpenoids from Laggera pterodonta. J. Asian. Nat. Prod. Res. 2007, 9, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Sy, L.K.; Brown, G.D.; Haynes, R. A Novel Endoperoxide and Related Sesquiterpenes from Artemisia Annua Which Are Possibly Derived from Ailylic Hydroperoxides. Tetrahedron 1998, 54, 4345–4356. [Google Scholar] [CrossRef]








| NO | 1 | 2 | ||
|---|---|---|---|---|
| δC Type | δH (J in Hz) | δC Type | δH (J in Hz) | |
| 1 | 41.3, CH | 1.75 (m) | 43.4, CH | 1.47 (d, 2.7) |
| 2 | 26.7, CH2 | 1.35 (d, 3.8), 1.88 (m) | 22.5, CH2 | 1.56 (ddd, 8.4, 3.7, 2.1), 1.95 (m) |
| 3 | 30.4, CH2 | 2.24 (m), 2.44 (tdt, 13.6, 5.3, 2.0) | 31.7, CH2 | 2.10 (s), 2.11 (d, 3.0) |
| 4 | 148.0, C | 144.1, C | ||
| 5 | 76.9, CH | 3.60 (s) | 121.6, CH | 5.44 (q, 1.6) |
| 6 | 87.9, C | 87.4, C | ||
| 7 | 42.4, CH | 3.21 (ddd, 10.5, 5.0, 3.4) | 77.2, C | |
| 8 | 30.1, CH2 | 1.29 (m), 1.95 (m) | 37.0, CH2 | 1.66 (m), 1.81 (m) |
| 9 | 32.1, CH2 | 1.16 (m), 1.61 (m) | 30.4, CH2 | 1.35 (dd, 7.6, 3.8), 1.52 (m) |
| 10 | 31.1, CH | 1.33 (m) | 30.8, CH | 1.50 (s) |
| 11 | 143.7, C | 147.6, C | ||
| 12 | 172.0, C | 171.0, C | ||
| 13 | 121.6, CH2 | 5.67 (d, 1.3), 6.09 (d, 1.5) | 119.9, CH2 | 5.70 (s), 6.12 (s) |
| 14 | 20.4, CH3 | 0.97 (d, 6.6) | 19.8, CH3 | 1.00 (d, 5.9) |
| 15 | 113.0, CH2 | 4.79 (t, 1.9), 4.86 (t, 2.1) | 23.5, CH3 | 1.71 (d, 1.3) |
| Compounds | HCT116 | CT26 |
|---|---|---|
| 2 | 20.0 | 49.9 |
| 5 | 16.7 | 14.9 |
| 16 | 21.5 | 53.5 |
| CPT11 | 24.4 | 44.3 |
| Gene | Forward Primer | Reverse Primer |
|---|---|---|
| CDK4 | TTTTGAGCATCCCAATGTTGTC | TCGACGAAACATCTCTTGATCT |
| CDK1 | CACAAAACTACAGGTCAAGTGG | GAGAAATTTCCCGAATTGCAGT |
| CDK6 | CGAACAGACAGAGAAACCAAAC | CTCGGTGTGAATGAAGAAAGTC |
| CCND1 | GTCCTACTTCAAATGTGTGCAG | GGGATGGTCTCCTTCATCTTAG |
| CCNB1 | GACTTTGCTTTTGTGACTGACA | CCCAGACCAAAGTTTAAAGCTC |
| β-actin | CAGCCTTCCTTCTTGGGTAT | TGGCATAGAGGTCTTACGG |
| Caspase3 | CACATGAUGCTUCCGACUGA | ATGGTTCGGTTACTACGGTCA |
| BCL2 | GACTTCGCCGAGATGTCCAG | GAACTCAAAGAAGGCCACAATC |
| Bax | CGAACTGGACAGTAACATGGAG | CAGTTTGCTGGCAAAGTAGAAA |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, X.; Chai, Y.; Lv, C.; Chen, Q.; Liu, J.; Wang, Y.; Chou, G. Sesquiterpenes from Artemisia annua and Their Cytotoxic Activities. Molecules 2022, 27, 5079. https://doi.org/10.3390/molecules27165079
Han X, Chai Y, Lv C, Chen Q, Liu J, Wang Y, Chou G. Sesquiterpenes from Artemisia annua and Their Cytotoxic Activities. Molecules. 2022; 27(16):5079. https://doi.org/10.3390/molecules27165079
Chicago/Turabian StyleHan, Xiao, Yao Chai, Cheng Lv, Qianqian Chen, Jinling Liu, Yongli Wang, and Guixin Chou. 2022. "Sesquiterpenes from Artemisia annua and Their Cytotoxic Activities" Molecules 27, no. 16: 5079. https://doi.org/10.3390/molecules27165079
APA StyleHan, X., Chai, Y., Lv, C., Chen, Q., Liu, J., Wang, Y., & Chou, G. (2022). Sesquiterpenes from Artemisia annua and Their Cytotoxic Activities. Molecules, 27(16), 5079. https://doi.org/10.3390/molecules27165079