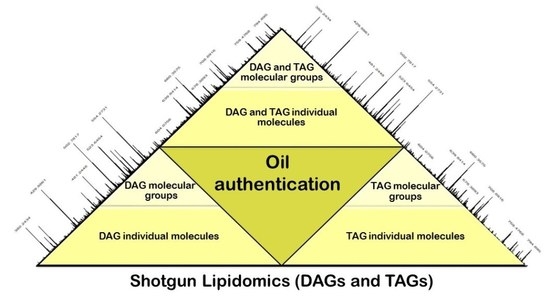
Shotgun Lipidomic Analysis for Differentiation of Niche Cold Pressed Oils
Abstract
:1. Introduction
2. Materials and Methods
2.1. Samples
2.2. Lipids Analysis
2.3. Multivariate Data Analysis
2.4. Results
2.4.1. Characteristics of DAG and TAG Profiles of Oils
2.4.2. Multivariate Data Analysis to Differentiate CA, FL, and HP Oils
3. Discussion
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Kostik, V.; Memeti, S.; Bauer, B. Fatty acid composition of edible oils and fats. J. Hyg. Eng. Des. 2013, 4, 112–116. [Google Scholar]
- Dhifi, W.; Bellili, S.; Jazi, S.; Bahloul, N.; Mnif, W. Essential Oils’ Chemical Characterization and Investigation of Some Biological Activities: A Critical Review. Medicines 2016, 3, 25. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campos, D.A.; Gómez-García, R.; Vilas-Boas, A.A.; Madureira, A.R.; Pintado, M.M. Management of fruit industrial by-products—A case study on circular economy approach. Molecules 2020, 25, 320. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Frankel, E.N. Chemistry of extra virgin olive oil: Adulteration, oxidative stability, and antioxidants. J. Agric. Food Chem. 2010, 58, 5991–6006. [Google Scholar] [CrossRef] [PubMed]
- Ben-Ayed, R.; Kamoun-Grati, N.; Rebai, A. An overview of the authentication of olive tree and oil. Compr. Rev. Food Sci. Food Saf. 2013, 12, 218–227. [Google Scholar] [CrossRef]
- Cavin, C.; Cottenet, G.; Fuerer, C.; Tran, L.A.; Zbinden, P. Food fraud vulnerabilities in the supply chain: An industry perspective. In Encyclopedia of Food Chemistry; Melton, L., Shahidi, F., Varelis, P., Eds.; Academic Press: Cambridge, MA, USA, 2019; Volume 1, pp. 670–678. [Google Scholar]
- Yan, J.; Erasmus, S.W.; Aguilera Toro, M.; Huang, H.; van Ruth, S.M. Food fraud: Assessing fraud vulnerability in the extra virgin olive oil supply chain. Food Control 2020, 111, 107081. [Google Scholar] [CrossRef]
- MacMahon, S. Contaminants in Food Lipids. In Woodhead Publishing Series in Food Science, Technology and Nutrition, Functional Dietary Lipids; Sanders, T.A.B., Ed.; Woodhead Publishing: Sawston, UK, 2016; pp. 195–222. [Google Scholar]
- Lerma-García, M.J.; Ramis-Ramos, G.; Herrero-Martínez, J.M.; Simó-Alfonso, E.F. Authentication of extra virgin olive oils by Fourier-transform infrared spectroscopy. Food Chem. 2010, 118, 78–83. [Google Scholar] [CrossRef]
- Alves, J.O.; Botelho, B.G.; Sena, M.M.; Augusti, R. Electrospray ionization mass spectrometry and partial least squares discriminant analysis applied to the quality control of olive oil. J. Mass Spectrom. 2013, 48, 1109–1115. [Google Scholar] [CrossRef] [PubMed]
- Carranco, N.; Farrés-Cebrián, M.; Saurina, J.; Núñez, O. Authentication and quantitation of fraud in extra virgin olive oils based on HPLC-UV fingerprinting and multivariate calibration. Foods 2018, 7, 44. [Google Scholar] [CrossRef] [Green Version]
- Gurdeniz, G.; Ozen, B. Detection of adulteration of extra-virgin olive oil by chemometric analysis of mid-infrared spectral data. Food Chem. 2009, 116, 519–525. [Google Scholar] [CrossRef] [Green Version]
- Xu, Z.; Morris, R.H.; Bencsik, M.; Newton, M.I. Detection of virgin olive oil adulteration using low field unilateral NMR. Sensors 2014, 14, 2028–2035. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tomaszewska-Gras, J. Rapid quantitative determination of butter adulteration with palm oil using the DSC technique. Food Control 2016, 60, 629–635. [Google Scholar] [CrossRef]
- Aparicio, R.; Aparicio-Ruíz, R. Authentication of vegetable oils by chromatographic techniques. J. Chromatogr. A 2000, 881, 93–104. [Google Scholar] [CrossRef]
- Marriott, P.J.; Shellie, R.; Cornwell, C. Gas chromatographic technologies for the analysis of essential oils. J. Chromatogr. A 2001, 936, 1–22. [Google Scholar] [CrossRef]
- Xu, B.; Zhang, L.; Wang, H.; Luo, D.; Li, P. Characterization and authentication of four important edible oils using free phytosterol profiles established by GC-GC-TOF/MS. Anal. Methods 2014, 6, 6860–6870. [Google Scholar] [CrossRef]
- Hu, N.; Wei, F.; Lv, X.; Wu, L.; Dong, X.Y.; Chen, H. Profiling of triacylglycerols in plant oils by high-performance liquid chromatography-atmosphere pressure chemical ionization mass spectrometry using a novel mixed-mode column. J. Chromatogr. B Anal. Technol. Biomed. Life Sci. 2014, 972, 65–72. [Google Scholar] [CrossRef] [PubMed]
- Lísa, M.; Holčapek, M. Characterization of triacylglycerol enantiomers using chiral HPLC/APCI-MS and synthesis of enantiomeric triacylglycerols. Anal. Chem. 2013, 85, 1852–1859. [Google Scholar] [CrossRef] [PubMed]
- Fauconnot, L.; Hau, J.; Aeschlimann, J.M.; Fay, L.B.; Dionisi, F. Quantitative analysis of triacylglycerol regioisomers in fats and oils using reversed-phase high-performance liquid chromatography and atmospheric pressure chemical ionization mass spectrometry. Rapid Commun. Mass Spectrom. 2004, 18, 218–224. [Google Scholar] [CrossRef] [PubMed]
- Mondello, L.; Tranchida, P.Q.; Stanek, V.; Jandera, P.; Dugo, G.; Dugo, P. Silver-ion reversed-phase comprehensive two-dimensional liquid chromatography combined with mass spectrometric detection in lipidic food analysis. J. Chromatogr. A 2005, 1086, 91–98. [Google Scholar] [CrossRef]
- Palyzová, A.; Řezanka, T. Separation of triacylglycerols containing allenic and acetylenic fatty acids by enantiomeric liquid chromatography-mass spectrometry. J. Chromatogr. A 2020, 1623, 461161. [Google Scholar] [CrossRef]
- Wei, F.; Hu, N.; Lv, X.; Dong, X.Y.; Chen, H. Quantitation of triacylglycerols in edible oils by off-line comprehensive two-dimensional liquid chromatography-atmospheric pressure chemical ionization mass spectrometry using a single column. J. Chromatogr. A 2015, 1404, 60–71. [Google Scholar] [CrossRef] [PubMed]
- Itabashi, Y.; Kuksis, A. Chiral High Performance Liquid Chromatography of Neutral Glycerolipids. In Encyclopedia of Lipidomics; Wenk, M., Ed.; Springer: Dordrecht, Germany, 2016; pp. 1–14. [Google Scholar]
- Rahmati, S.; Julkapli, N.M.; Yehye, W.A.; Basirun, W.J. Identification of meat origin in food products—A review. Food Control 2016, 68, 379–390. [Google Scholar] [CrossRef]
- Debode, F.; Janssen, E.; Marien, A.; Berben, G. DNA detection by conventional and real-time PCR after extraction from vegetable oils. JAOCS J. Am. Oil Chem. Soc. 2012, 89, 1249–1257. [Google Scholar] [CrossRef]
- Malik, A.K.; Blasco, C.; Picó, Y. Liquid chromatography-mass spectrometry in food safety. J. Chromatogr. A 2010, 1217, 4018–4040. [Google Scholar] [CrossRef] [PubMed]
- Stachniuk, A.; Sumara, A.; Montowska, M.; Fornal, E. Liquid chromatography–mass spectrometry bottom-up proteomic methods in animal species analysis of processed meat for food authentication and the detection of adulterations. Mass Spectrom. Rev. 2019, 40, 3–30. [Google Scholar] [CrossRef] [PubMed]
- Nagy, K.; Bongiorno, D.; Avellone, G.; Agozzino, P.; Ceraulo, L.; Vékey, K. High performance liquid chromatography-mass spectrometry based chemometric characterization of olive oils. J. Chromatogr. A 2005, 1078, 90–97. [Google Scholar] [CrossRef] [PubMed]
- Jakab, A.; Héberger, K.; Forgács, E. Comparative analysis of different plant oils by high-performance liquid chromatography-atmospheric pressure chemical ionization mass spectrometry. J. Chromatogr. A 2002, 976, 255–263. [Google Scholar] [CrossRef]
- Siddabasave, S.G.; Minami, Y.; Gowda, D.; Furuko, D.; Chiba, H.; Hui, S.P. Lipidomic analysis of non-esterified furan fatty acids and fatty acid compositions in dietary shellfish and salmon by UHPLC/LTQ-Orbitrap-MS. Food Res. Int. 2021, 144, 110325. [Google Scholar]
- Han, X.; Cheng, H. Characterization and direct quantitation of cerebroside molecular species from lipid extracts by shotgun lipidomics. J. Lipid Res. 2005, 17, 264–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, X.; Yang, K.; Yang, J.; Fikes, K.N.; Cheng, H.; Gross, R.W. Factors influencing the electrospray intrasource separation and selective ionization of glycerophospholipids. J. Am. Soc. Mass Spectrom. 2006, 17, 264–274. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, K.; Han, X. Accurate quantification of lipid species by electrospray ionization mass spectrometry—Meets a key challenge in lipidomics. Metabolites 2011, 1, 21–40. [Google Scholar] [CrossRef] [PubMed]
- Paglia, G.; Kliman, M.; Claude, E.; Geromanos, S.; Astarita, G. Applications of ion-mobility mass spectrometry for lipid analysis. Anal. Bioanal. Chem. 2015, 407, 4995–5007. [Google Scholar] [CrossRef] [PubMed]
- Surma, M.A.; Herzog, R.; Vasilj, A.; Klose, C.; Christinat, N.; Morin-Rivron, D.; Simons, K.; Masoodi, M.; Sampaio, J.L. An automated shotgun lipidomics platform for high throughput, comprehensive, and quantitative analysis of blood plasma intact lipids. Eur. J. Lipid Sci. Technol. 2015, 117, 1540–1549. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nielsen, I.Ø.; Maeda, K.; Bilgin, M. Global Monitoring of the Mammalian Lipidome by Quantitative Shotgun Lipidomics. Methods Mol. Biol. 2017, 1609, 123–139. [Google Scholar] [PubMed]
- Fu, T.; Knittelfelder, O.; Geffard, O.; Clément, Y.; Testet, E.; Elie, N.; Touboul, D.; Abbaci, K.; Shevchenko, A.; Lemoine, J.; et al. Shotgun lipidomics and mass spectrometry imaging unveil diversity and dynamics in Gammarus fossarum lipid composition. iScience 2021, 24, 102115. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Wang, C.; Han, R.H.; Han, X. Novel advances in shotgun lipidomics for biology and medicine. Prog. Lipid Res. 2016, 61, 83–108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shen, Q.; Wang, Y.; Gong, L.; Guo, R.; Dong, W.; Cheung, H.Y. Shotgun lipidomics strategy for fast analysis of phospholipids in fisheries waste and its potential in species differentiation. J. Agric. Food Chem. 2012, 60, 9384–9393. [Google Scholar] [CrossRef] [PubMed]
- Capriotti, A.L.; Cerrato, A.; Aita, S.E.; Montone, C.M.; Piovesana, S.; Laganà, A.; Cavaliere, C. Degradation of the polar lipid and fatty acid molecular species in extra virgin olive oil during storage based on shotgun lipidomics. J. Chromatogr. A 2021, 1639, 461881. [Google Scholar] [CrossRef] [PubMed]
- Herzog, R.; Schwudke, D.; Schuhmann, K.; Sampaio, J.L.; Bornstein, S.R.; Schroeder, M.; Shevchenko, A. A novel informatics concept for high-throughput shotgun lipidomics based on the molecular fragmentation query language. Genome Biol. 2011, 12, R8. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Schrimpe-Rutledge, A.C.; Codreanu, S.G.; Sherrod, S.D.; McLean, J.A. Untargeted Metabolomics Strategies—Challenges and Emerging Directions. J. Am. Soc. Mass Spectrom. 2016, 27, 1897–1905. [Google Scholar] [CrossRef] [Green Version]
- Szabó, D.; Schlosser, G.; Vékey, K.; Drahos, L.; Révész, Á. Collision energies on QTof and Orbitrap instruments: How to make proteomics measurements comparable? J. Mass Spectrom. 2021, 56, e4693. [Google Scholar] [CrossRef] [PubMed]



| Lipid Class | Number of Groups/Individuals | ||||
|---|---|---|---|---|---|
| Camelina | Flax | Hemp | Total | ||
| Molecular groups (species) | DAG | 11 | 10 | 11 | 13 |
| TAG | 65 | 52 | 48 | 67 | |
| Individual molecules (subspecies) | DAG | 12 | 13 | 15 | 18 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Nikolaichuk, H.; Przykaza, K.; Kozub, A.; Montowska, M.; Wójcicka, G.; Tomaszewska-Gras, J.; Fornal, E. Shotgun Lipidomic Analysis for Differentiation of Niche Cold Pressed Oils. Molecules 2022, 27, 1848. https://doi.org/10.3390/molecules27061848
Nikolaichuk H, Przykaza K, Kozub A, Montowska M, Wójcicka G, Tomaszewska-Gras J, Fornal E. Shotgun Lipidomic Analysis for Differentiation of Niche Cold Pressed Oils. Molecules. 2022; 27(6):1848. https://doi.org/10.3390/molecules27061848
Chicago/Turabian StyleNikolaichuk, Hanna, Kacper Przykaza, Anna Kozub, Magdalena Montowska, Grażyna Wójcicka, Jolanta Tomaszewska-Gras, and Emilia Fornal. 2022. "Shotgun Lipidomic Analysis for Differentiation of Niche Cold Pressed Oils" Molecules 27, no. 6: 1848. https://doi.org/10.3390/molecules27061848
APA StyleNikolaichuk, H., Przykaza, K., Kozub, A., Montowska, M., Wójcicka, G., Tomaszewska-Gras, J., & Fornal, E. (2022). Shotgun Lipidomic Analysis for Differentiation of Niche Cold Pressed Oils. Molecules, 27(6), 1848. https://doi.org/10.3390/molecules27061848