In Silico Analysis of Nanoplastics’ and β-amyloid Fibrils’ Interactions
Abstract
:1. Introduction
2. Results and discussion
2.1. Nanoplastics Models
2.2. Aβ Fibril Model
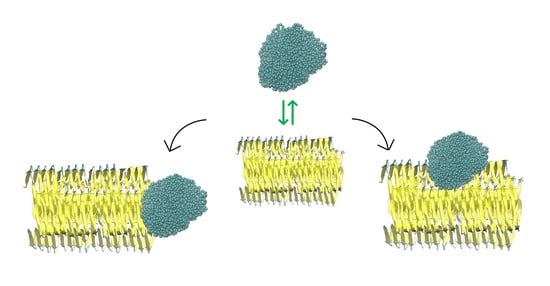
2.3. NP–Aβ Fibril Interactions
3. Conclusions
4. Materials and Methods
4.1. Aβ Fibril Model
4.2. Nanoplastics Models
4.3. MD Simulations of Fibril–NP Complexes
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Prüst, M.; Meijer, J.; Westerink, R.H.S. The Plastic Brain: Neurotoxicity of Micro- and Nanoplastics. Part. Fibre Toxicol. 2020, 17, 1–16. [Google Scholar] [CrossRef] [PubMed]
- Kihara, S.; Köper, I.; Mata, J.P.; McGillivray, D.J. Reviewing Nanoplastic Toxicology: It’s an Interface Problem. Adv. Colloid Interface Sci. 2021, 288, 102337. [Google Scholar] [CrossRef] [PubMed]
- Marcelino, R.C.; Cardoso, R.M.; Domingues, E.L.B.C.; Gonçalves, R.V.; Lima, G.D.A.; Novaes, R.D. The Emerging Risk of Microplastics and Nanoplastics on the Microstructure and Function of Reproductive Organs in Mammals: A Systematic Review of Preclinical Evidence. Life Sci. 2022, 295, 1–9. [Google Scholar] [CrossRef] [PubMed]
- Revel, M.; Châtel, A.; Mouneyrac, C. Micro(Nano)Plastics: A Threat to Human Health? Curr. Opin. Environ. Sci. Health 2018, 1, 17–23. [Google Scholar] [CrossRef]
- Jenner, L.C.; Rotchell, J.M.; Bennett, R.T.; Cowen, M.; Tentzeris, V.; Sadofsky, L.R. Detection of Microplastics in Human Lung Tissue Using ΜFTIR Spectroscopy. Sci. Total Environ. 2022, 831, 154907. [Google Scholar] [CrossRef] [PubMed]
- Kik, K.; Bukowska, B.; Sicińska, P. Polystyrene Nanoparticles: Sources, Occurrence in the Environment, Distribution in Tissues, Accumulation and Toxicity to Various Organisms. Environ. Pollut. 2020, 262, 114297. [Google Scholar] [CrossRef]
- Leslie, H.A.; van Velzen, M.J.M.; Brandsma, S.H.; Vethaak, A.D.; Garcia-Vallejo, J.J.; Lamoree, M.H. Discovery and Quantification of Plastic Particle Pollution in Human Blood. Environ. Int. 2022, 163, 107199. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, S.; Duan, Z.; Wang, L. Pulmonary Toxicology Assessment of Polyethylene Terephthalate Nanoplastic Particles in Vitro. Environ. Int. 2022, 162, 107177. [Google Scholar] [CrossRef]
- Järvenpää, J.; Perkkiö, M.; Laitinen, R.; Lahtela-Kakkonen, M. PE and PET Oligomers’ Interplay with Membrane Bilayers. Sci. Rep. 2022, 12, 2234. [Google Scholar] [CrossRef]
- Liu, L.; Xu, K.; Zhang, B.; Ye, Y.; Zhang, Q.; Jiang, W. Cellular Internalization and Release of Polystyrene Microplastics and Nanoplastics. Sci. Total Environ. 2021, 779, 146523. [Google Scholar] [CrossRef]
- Hollóczki, O.; Gehrke, S. Nanoplastics Can Change the Secondary Structure of Proteins. Sci. Rep. 2019, 9, 16013. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fleischer, C.C.; Payne, C.K. Nanoparticle-Cell Interactions: Molecular Structure of the Protein Corona and Cellular Outcomes. Acc. Chem. Res. 2014, 47, 2651–2659. [Google Scholar] [CrossRef] [PubMed]
- Hollóczki, O. Evidence for Protein Misfolding in the Presence of Nanoplastics. Int. J. Quantum Chem. 2021, 121, e26372. [Google Scholar] [CrossRef]
- Nowak, M.; Brown, T.D.; Graham, A.; Helgeson, M.E.; Mitragotri, S. Size, Shape, and Flexibility Influence Nanoparticle Transport across Brain Endothelium under Flow. Bioeng. Transl. Med. 2020, 5, e10153. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Windheim, J.; Colombo, L.; Battajni, N.C.; Russo, L.; Cagnotto, A.; Diomede, L.; Bigini, P.; Vismara, E.; Fiumara, F.; Gabbrielli, S.; et al. Micro- and Nanoplastics ’ Effects on Protein Folding and Amyloidosis. Int. J. Mol. Sci. 2022, 23, 10329. [Google Scholar] [CrossRef] [PubMed]
- Morgan, C.; Colombres, M.; Nuñez, M.T.; Inestrosa, N.C. Structure and Function of Amyloid in Alzheimer’s Disease. Prog. Neurobiol. 2004, 74, 323–349. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Walter, J. Phosphorylation of Amyloid Beta Peptides. Aging 2011, 3, 803–812. [Google Scholar] [CrossRef] [PubMed]
- Mahmoudi, M.; Kalhor, H.R.; Laurent, S.; Lynch, I. Protein Fibrillation and Nanoparticle Interactions: Opportunities and Challenges. Nanoscale 2013, 5, 2570–2588. [Google Scholar] [CrossRef]
- Linse, S.; Cabaleiro-Lago, C.; Xue, W.F.; Lynch, I.; Lindman, S.; Thulin, E.; Radford, S.E.; Dawson, K.A. Nucleation of Protein Fibrillation by Nanoparticles. Proc. Natl. Acad. Sci. USA 2007, 104, 8691–8696. [Google Scholar] [CrossRef] [Green Version]
- Cabaleiro-Lago, C.; Quinlan-Pluck, F.; Lynch, I.; Lindman, S.; Minogue, A.M.; Thulin, E.; Walsh, D.M.; Dawson, K.A.; Linse, S. Inhibition of Amyloid β Protein Fibrillation by Polymeric Nanoparticles. J. Am. Chem. Soc. 2008, 130, 15437–15443. [Google Scholar] [CrossRef]
- Cabaleiro-Lago, C.; Quinlan-Pluck, F.; Lynch, I.; Dawson, K.A.; Linse, S. Dual Effect of Amino Modified Polystyrene Nanoparticles on Amyloid β Protein Fibrillation. ACS Chem. Neurosci. 2010, 1, 279–287. [Google Scholar] [CrossRef]
- Calvaresi, M.; Hoefinger, S.; Zerbetto, F. Probing the Structure of Lysozyme-Carbon-Nanotube Hybrids with Molecular Dynamics. Chem. —Eur. J. 2012, 18, 4308–4313. [Google Scholar] [CrossRef] [PubMed]
- Liu, G.R.; Cheng, Y.; Mi, D.; Li, Z.R. A Study on Self-Insertion of Peptides into Single-Walled Carbon Nanotubes Based on Molecular Dynamics Simulation. Int. J. Mod. Phys. C 2005, 16, 1239–1250. [Google Scholar] [CrossRef]
- Sun, X.; Feng, Z.; Hou, T.; Li, Y. Mechanism of Graphene Oxide as an Enzyme Inhibitor from Molecular Dynamics Simulations. ACS Appl. Mater. Interfaces 2014, 6, 7153–7163. [Google Scholar] [CrossRef] [PubMed]
- Baweja, L.; Balamurugan, K.; Subramanian, V.; Dhawan, A. Effect of Graphene Oxide on the Conformational Transitions of Amyloid Beta Peptide: A Molecular Dynamics Simulation Study. J. Mol. Graph. Model. 2015, 61, 175–185. [Google Scholar] [CrossRef]
- Fröhlich, E. The Role of Surface Charge in Cellular Uptake and Cytotoxicity of Medical Nanoparticles. Int. J. Nanomed. 2012, 7, 5577–5591. [Google Scholar] [CrossRef] [Green Version]
- Berendsen, H.J.C.; van der Spoel, D.; van Drunen, R. GROMACS: A Message-Passing Parallel Molecular Dynamics Implementation. Comput. Phys. Commun. 1995, 91, 43–56. [Google Scholar] [CrossRef]
- Marrink, S.J.; Risselada, H.J.; Yefimov, S.; Tieleman, D.P.; de Vries, A.H. The MARTINI Force Field: Coarse Grained Model for Biomolecular Simulations. J. Phys. Chem. B 2007, 111, 7812–7824. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Monticelli, L.; Kandasamy, S.K.; Periole, X.; Larson, R.G.; Tieleman, D.P.; Marrink, S.-J. The MARTINI Coarse-Grained Force Field: Extension to Proteins. J. Chem. Theory Comput. 2008, 4, 819–834. [Google Scholar] [CrossRef]
- Gremer, L.; Schölzel, D.; Schenk, C.; Reinartz, E.; Labahn, J.; Ravelli, R.B.G.; Tusche, M.; Lopez-Iglesias, C.; Hoyer, W.; Heise, H.; et al. Fibril Structure of Amyloid-β(1–42) by Cryo–Electron Microscopy. Science 2017, 358, 116–119. [Google Scholar] [CrossRef]
- Phillips, J.C.; Braun, R.; Wang, W.; Gumbart, J.; Tajkhorshid, E.; Villa, E.; Chipot, C.; Skeel, R.D.; Kalé, L.; Schulten, K. Scalable Molecular Dynamics with NAMD. J. Comput. Chem. 2005, 26, 1781–1802. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Panizon, E.; Bochicchio, D.; Monticelli, L.; Rossi, G. MARTINI Coarse-Grained Models of Polyethylene and Polypropylene. J. Phys. Chem. B 2015, 119, 8209–8216. [Google Scholar] [CrossRef] [PubMed]
- Rossi, G.; Monticelli, L.; Puisto, S.R.; Vattulainen, I.; Ala-Nissila, T. Coarse-Graining Polymers with the MARTINI Force-Field: Polystyrene as a Benchmark Case. Soft Matter 2011, 7, 698–708. [Google Scholar] [CrossRef] [Green Version]
- Gautieri, A.; Ionita, M.; Silvestri, D.; Votta, E.; Vesentini, S.; Fiore, G.B.; Barbani, N.; Ciardelli, G.; Redaelli, A. Computer-Aided Molecular Modeling and Experimental Validation of Water Permeability Properties in Biosynthetic Materials. J. Comput. Theor. Nanosci. 2010, 7, 1287–1293. [Google Scholar] [CrossRef]
- Gautieri, A.; Beeg, M.; Gobbi, M.; Rigoldi, F.; Colombo, L.; Salmona, M. The Anti-Amyloidogenic Action of Doxycycline: A Molecular Dynamics Study on the Interaction with Aβ42. Int. J. Mol. Sci. 2019, 20, 4641. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rigoldi, F.; Donini, S.; Giacomina, F.; Sorana, F.; Redaelli, A.; Bandiera, T.; Parisini, E.; Gautieri, A. Thermal Stabilization of the Deglycating Enzyme Amadoriase i by Rational Design. Sci. Rep. 2018, 8, 3042. [Google Scholar] [CrossRef]








| PS | PE | PP | PS4+ | PS10+ | PS20+ | PS4- | PS10- | PS20- | |
|---|---|---|---|---|---|---|---|---|---|
| System 1 | side | frontal | side | - | side | side | - | - | - |
| System 2 | - | side | frontal | frontal | frontal | - | - | - | - |
| System 3 | side | - | frontal | side | - | side | - | - | - |
| System 4 | side | side | - | side | side | side | - | - | - |
| System 5 | side | frontal | frontal | side | side | side | - | - | - |
| System 6 | - | side | frontal | side | - | side | - | - | - |
| System 7 | side | side | side | frontal | side | side | - | - | - |
| System 8 | side | frontal | frontal | - | frontal | side | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Gabbrielli, S.; Colnaghi, L.; Mazzuoli-Weber, G.; Redaelli, A.C.L.; Gautieri, A. In Silico Analysis of Nanoplastics’ and β-amyloid Fibrils’ Interactions. Molecules 2023, 28, 388. https://doi.org/10.3390/molecules28010388
Gabbrielli S, Colnaghi L, Mazzuoli-Weber G, Redaelli ACL, Gautieri A. In Silico Analysis of Nanoplastics’ and β-amyloid Fibrils’ Interactions. Molecules. 2023; 28(1):388. https://doi.org/10.3390/molecules28010388
Chicago/Turabian StyleGabbrielli, Silvia, Luca Colnaghi, Gemma Mazzuoli-Weber, Alberto Cesare Luigi Redaelli, and Alfonso Gautieri. 2023. "In Silico Analysis of Nanoplastics’ and β-amyloid Fibrils’ Interactions" Molecules 28, no. 1: 388. https://doi.org/10.3390/molecules28010388
APA StyleGabbrielli, S., Colnaghi, L., Mazzuoli-Weber, G., Redaelli, A. C. L., & Gautieri, A. (2023). In Silico Analysis of Nanoplastics’ and β-amyloid Fibrils’ Interactions. Molecules, 28(1), 388. https://doi.org/10.3390/molecules28010388