Identification of Turtle-Shell Growth Year Using Hyperspectral Imaging Combined with an Enhanced Spatial–Spectral Attention 3DCNN and a Transformer
Abstract
:1. Introduction
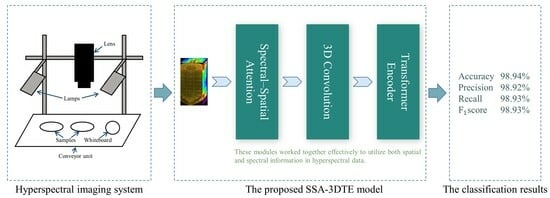
- First, inspired by the study in [28], a spectral–spatial attention mechanism module is developed to selectively extract the low-level features of the neighborhood pixel blocks and reduce spectral redundant information in raw 3D HSI data. Generally, extracting only spectral data from a region of interest (ROI) as a one-dimension vector could lead to the loss of external spatial information, or spectral and image information of HSI data could be considered separately [29]. Also, this wealth of spatial and spectral information of 3D hyperspectral images unequally contributes to the final classification, particularly considering the correlation and redundancy within the spectral spectrum. To deal with the aforementioned challenges, the proposed model utilizes a sequential stacking of the spectral–spatial attention (SSA) module to refine the learned joint spectral–spatial features.
- Second, regarding the extracted 3D feature map, three-dimensional convolution and the transformer are employed to effectively capture the local and global information for optimizing the classification process. Joint spatial–spectral feature representation is facilitated by 3D convolution. Moreover, taking into account that HSI can be perceived as sequence data, the convolution operation is confined by its receptive field, hence it cannot maintain a balance between model performance and depth [30]. To overcome these limitations and enhance the efficacy of spectral–spatial feature extraction, a novel approach was adopted. This approach integrated a transformer encoder block with multi-head self-attention (MSA) mechanisms, called the TE block, which can effectively address the long-distance dependencies inherent in the spectral band information of the hyperspectral image data.
2. Results and Discussion
2.1. Spectral Profile
2.2. Parameter Analysis
- (1)
- Principal component analysis: PCA was utilized to process the HSI data in order to mitigate the computational burden and spectral dimensionality. Here, the principal component numbers were evaluated as 20, 30, 40, 50, 60, 70, 80, 90, and 100. It can be seen from Figure 2a that the principal component numbers have an impact on the classification performance. Among them, the worst classification accuracy is 94.73% when the number of principal components is 20, and the highest is with 60 components. The main reason is that regarding the principal component number, if the setting is too small, most of the valid features will be rejected, and if the setting is too large, it may contain some redundant spectral information, also with an increased computational burden. Also, the model with 60 principal components maintains a smaller variance, which means that it obtains a relatively stable performance. For the subsequent trials, the principal component numbers are set to 60.
- (2)
- Learning rate: To ensure effective training, selecting an appropriate learning rate is essential as it greatly affects the gradient descent rate of the model and influences the convergence performance and speed of the model. In this study, an analysis of various learning rates was conducted, including 0.0005, 0.001, 0.003, 0.005, 0.01, and 0.03. Figure 2b shows that an appropriate increase in the learning rate has a positive effect on the model performance, and the effect reaches an optimal value for accuracy with a learning rate of 0.005, but a further increase will cause a significant decrease in accuracy. Based on the abovementioned results, the learning rate is set to 0.005 in the following experiments.
- (3)
- Number of heads in transformer block: The number of heads in the TE block is varied, with the head cardinality set to 2, 4, 8, and 16. Generally, an appropriate increase in the number of SA heads should enable the model to learn richer and more robust features. As the number of SA heads increases, the classification accuracy increases, but this increase comes at the cost of an increase in total network parameters, which can make network training more difficult and ultimately reduce its classification accuracy. Figure 2c shows that when the number of SA heads is equal to 4, the classification accuracy reaches the maximum value.
- (4)
- Number of 3D convolution kernels: The influences of the numbers of 3D convolution kernels on the accuracy are illustrated in Figure 2d. The results show that the classification increased first and then decreased with more 3D kernels, and it peaks at 16 3D kernels. Overall, Figure 2d suggests that the classification accuracy is not significantly affected by the number of convolution kernels, indicating the stability of the model’s performance. Among them, the model with 16 kernels achieved the best performance.
2.3. Ablation Experiments
2.4. Comparative Performance of Various Methods
2.4.1. Discrimination Results of Representative Models Using Only Spectral Information
2.4.2. Comparing with Representative Deep Learning-Based Methods
2.5. Confusion Matrix of Proposed Model
3. Materials and Preprocessing
3.1. Samples Preparation
3.2. Hyperspectral Imaging System and Image Acquisition
3.3. Hyperspectral Image Calibration
3.4. ROI Selection and Dimension Reduction
4. Methods
4.1. Spectral–Spatial Attention Block
4.1.1. Spectral Attention Module
4.1.2. Spatial Attention Module
4.2. 3D Convolution Block
4.3. Transformer Encoder Block
4.4. Overview of the Proposed Model
4.5. Experimental Settings
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- National Pharmacopoeia Committee. Pharmacopoeia of the People’s Republic of China; China Medical Science Press: Beijing, China, 2015; Volume 1, pp. 180–181. [Google Scholar]
- Li, L.; Cheung, H. Turtle shell extract as a functional food and its component-based comparison among different species. Hong Kong Pharm. J. 2012, 19, 33–37. [Google Scholar]
- Chen, M.; Han, X.; Ma, J.; Liu, X.; Li, M.; Lv, X.; Lu, H.; Ren, G.; Liu, C. Identification of Plastrum Testudinis used in traditional medicine with DNA mini-barcodes. Rev. Bras. De Farmacogn. 2018, 28, 267–272. [Google Scholar] [CrossRef]
- Zhao, Z.; Liang, Z.; Ping, G. Macroscopic identification of Chinese medicinal materials: Traditional experiences and modern understanding. J. Ethnopharmacol. 2011, 134, 556–564. [Google Scholar] [CrossRef]
- Jiang, Y.; David, B.; Tu, P.; Barbin, Y. Recent analytical approaches in quality control of traditional Chinese medicines—A review. Anal. Chim. Acta 2010, 657, 9–18. [Google Scholar] [CrossRef] [PubMed]
- Ristivojevi’c, P.M.; Tahir, A.; Malfent, F.; Opsenica, D.M.; Rollinger, J.M. High-performance thin-layer chromatography/bioautography and liquid chromatography-mass spectrometry hyphenated with chemometrics for the quality assessment of Morus alba samples. J. Chromatogr. A 2019, 1594, 190–198. [Google Scholar] [CrossRef]
- Luo, D.; Chen, J.; Gao, L.; Liu, Y.; Wu, J. Geographical origin identification and quality control of Chinese chrysanthemum flower teas using gas chromatography–mass spectrometry and olfactometry and electronic nose combined with principal component analysis. Int. J. Food Sci. Technol. 2017, 52, 714–723. [Google Scholar] [CrossRef]
- Liu, F.; Ma, N.; He, C.; Hu, Y.; Li, P.; Chen, M.; Su, H.; Wan, J. Qualitative and quantitative analysis of the saponins in Panax notoginseng leaves using ultra-performance liquid chromatography coupled with time-of-flight tandem mass spectrometry and high performance liquid chromatography coupled with UV detector. J. Ginseng Res. 2018, 42, 149–157. [Google Scholar] [CrossRef] [PubMed]
- Woodburn, D.B.; Kinsel, M.J.; Poll, C.P.; Langan, J.N.; Haman, K.; Gamble, K.C.; Maddox, C.; Jeon, A.B.; Wellehan, J.F.; Ossiboff, R.J.; et al. Shell lesions associated with Emydomyces testavorans infection in freshwater aquatic turtles. Vet. Pathol. 2021, 58, 578–586. [Google Scholar] [CrossRef] [PubMed]
- Yang, H.; Zheng, J.; Wang, H.; Li, N.; Yang, Y.; Shen, Y. Comparative proteomic analysis of three gelatinous chinese medicines and their authentications by tryptic-digested peptides profiling using matrix-assisted laser desorption/ionization-time of flight/time of flight mass spectrometry. Pharmacogn. Mag. 2017, 13, 663. [Google Scholar] [PubMed]
- Fazari, A.; Pellicer Valero, O.J.; Gómez Sanchıs, J.; Bernardi, B.; Cubero, S.; Benalia, S.; Zimbalatti, G.; Blasco, J. Application of deep convolutional neural networks for the detection of anthracnose in olives using VIS/NIR hyperspectral images. Comput. Electron. Agric. 2021, 187, 106252. [Google Scholar] [CrossRef]
- He, J.; He, Y.; Zhang, C. Determination and visualization of peimine and peiminine content in Fritillaria thunbergii bulbi treated by sulfur fumigation using hyperspectral imaging with chemometrics. Molecules 2017, 22, 1402. [Google Scholar] [CrossRef] [PubMed]
- Long, W.; Wang, S.; Suo, Y.; Chen, H.; Bai, X.; Yang, X.; Zhou, Y.; Yang, J.; Fu, H. Fast and non-destructive discriminating the geographical origin of Hangbaiju by hyperspectral imaging combined with chemometrics. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2023, 284, 121786. [Google Scholar] [CrossRef]
- Pan, Y.; Zhang, H.; Chen, Y.; Gong, X.; Yan, J.; Zhang, H. Applications of Hyperspectral Imaging Technology Combined with Machine Learning in Quality Control of Traditional Chinese Medicine from the Perspective of Artificial Intelligence: A Review. Crit. Rev. Anal. Chem. 2023, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ding, R.; Yu, L.; Wang, C.; Zhong, S.; Gu, R. Quality assessment of traditional Chinese medicine based on data fusion combined with machine learning: A review. Crit. Rev. Anal. Chem. 2023, 1–18. [Google Scholar] [CrossRef] [PubMed]
- Xu, Y.; Zhang, J.; Wang, Y. Recent trends of multi-source and non-destructive information for quality authentication of herbs and spices. Food Chem. 2023, 398, 133939. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Huang, H.; Zuo, Z.; Wang, Y. Comprehensive quality assessment of Dendrubium officinale using ATR-FTIR spectroscopy combined with random forest and support vector machine regression. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2018, 205, 637–648. [Google Scholar] [CrossRef]
- Ru, C.; Li, Z.; Tang, R. A hyperspectral imaging approach for classifying geographical origins of rhizoma atractylodis macro- cephalae using the fusion of spectrum-image in VNIR and SWIR ranges (VNIR-SWIR-FuSI). Sensors 2019, 19, 2045. [Google Scholar] [CrossRef]
- Han, Q.; Li, Y.; Yu, L. Classification of glycyrrhiza seeds by near infrared hyperspectral imaging technology. In Proceedings of the 2019 International Conference on High Performance Big Data and Intelligent Systems (HPBD&IS), Shenzhen, China, 9–11 May 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 141–145. [Google Scholar]
- Wang, L.; Li, J.; Qin, H.; Xu, J.; Zhang, X.; Huang, L. Selecting near-infrared hyperspectral wavelengths based on one-way ANOVA to identify the origin of Lycium barbarum. In Proceedings of the 2019 International Conference on High Performance Big Data and Intelligent Systems (HPBD&IS), Shenzhen, China, 9–11 May 2019; IEEE: Piscataway, NJ, USA, 2019; pp. 122–125. [Google Scholar]
- Yao, K.; Sun, J.; Tang, N.; Xu, M.; Cao, Y.; Fu, L.; Zhou, X.; Wu, X. Nondestructive detection for Panax notoginseng powder grades based on hyperspectral imaging technology combined with CARS-PCA and MPA-LSSVM. J. Food Process Eng. 2021, 44, e13718. [Google Scholar] [CrossRef]
- Xiaobo, Z.; Jiewen, Z.; Povey, M.J.; Holmes, M.; Hanpin, M. Variables selection methods in near-infrared spectroscopy. Anal. Chim. Acta 2010, 667, 14–32. [Google Scholar] [CrossRef]
- Zhong, Y.; Ru, C.; Wang, S.; Li, Z.; Cheng, Y. An online, non-destructive method for simultaneously detecting chemical, biological, and physical properties of herbal injections using hyperspectral imaging with artificial intelligence. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2022, 264, 120250. [Google Scholar] [CrossRef]
- Yan, T.; Duan, L.; Chen, X.; Gao, P.; Xu, W. Application and interpretation of deep learning methods for the geographical origin identification of Radix Glycyrrhizae using hyperspectral imaging. RSC Adv. 2020, 10, 41936–41945. [Google Scholar] [CrossRef] [PubMed]
- Kabir, M.H.; Guindo, M.L.; Chen, R.; Liu, F.; Luo, X.; Kong, W. Deep Learning Combined with Hyperspectral Imaging Technology for Variety Discrimination of Fritillaria thunbergii. Molecules 2022, 27, 6042. [Google Scholar] [CrossRef] [PubMed]
- Dong, F.; Hao, J.; Luo, R.; Zhang, Z.; Wang, S.; Wu, K.; Liu, M. Identification of the proximate geographical origin of wolfberries by two-dimensional correlation spectroscopy combined with deep learning. Comput. Electron. Agric. 2022, 198, 107027. [Google Scholar] [CrossRef]
- Mu, Q.; Kang, Z.; Guo, Y.; Chen, L.; Wang, S.; Zhao, Y. Hyperspectral image classification of wolfberry with different geographical origins based on three-dimensional convolutional neural network. Int. J. Food Prop. 2021, 24, 1705–1721. [Google Scholar] [CrossRef]
- Zhu, M.; Jiao, L.; Liu, F.; Yang, S.; Wang, J. Residual spectral–spatial attention network for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2020, 59, 449–462. [Google Scholar] [CrossRef]
- Liu, Y.; Zhou, S.; Han, W.; Liu, W.; Qiu, Z.; Li, C. Convolutional neural network for hyperspectral data analysis and effective wavelengths selection. Anal. Chim. Acta 2019, 1086, 46–54. [Google Scholar] [CrossRef] [PubMed]
- Qing, Y.; Liu, W.; Feng, L.; Gao, W. Improved transformer net for hyperspectral image classification. Remote Sens. 2021, 13, 2216. [Google Scholar] [CrossRef]
- Araújo, M.C.U.; Saldanha, T.C.B.; Galvao, R.K.H.; Yoneyama, T.; Chame, H.C.; Visani, V. The successive projections algorithm for variable selection in spectroscopic multicomponent analysis. Chemom. Intell. Lab. Syst. 2001, 57, 65–73. [Google Scholar] [CrossRef]
- Cai, W.; Li, Y.; Shao, X. A variable selection method based on uninformative variable elimination for multivariate calibration of near-infrared spectra. Chemom. Intell. Lab. Syst. 2008, 90, 188–194. [Google Scholar] [CrossRef]
- Li, H.; Liang, Y.; Xu, Q.; Cao, D. Key wavelengths screening using competitive adaptive reweighted sampling method for multivariate calibration. Anal. Chim. Acta 2009, 648, 77–84. [Google Scholar] [CrossRef]
- Wu, N.; Zhang, C.; Bai, X.; Du, X.; He, Y. Discrimination of Chrysanthemum varieties using hyperspectral imaging combined with a deep convolutional neural network. Molecules 2018, 23, 2831. [Google Scholar] [CrossRef] [PubMed]
- Hao, J.; Dong, F.; Li, Y.; Wang, S.; Cui, J.; Zhang, Z.; Wu, K. Investigation of the data fusion of spectral and textural data from hyperspectral imaging for the near geographical origin discrimination of wolfberries using 2D-CNN algorithms. Infrared Phys. Technol. 2022, 125, 104286. [Google Scholar] [CrossRef]
- Nagasubramanian, K.; Jones, S.; Singh, A.K.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B. Plant disease identification using explainable 3D deep learning on hyperspectral images. Plant Methods 2019, 15, 98. [Google Scholar] [CrossRef] [PubMed]
- Roy, S.K.; Krishna, G.; Dubey, S.R.; Chaudhuri, B.B. HybridSN: Exploring 3-D–2-D CNN feature hierarchy for hyperspectral image classification. IEEE Geosci. Remote Sens. Lett. 2019, 17, 277–281. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Las Vegas, NV, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Wang, L.; Peng, J.; Sun, W. Spatial–spectral squeeze-and-excitation residual network for hyperspectral image classification. Remote Sens. 2019, 11, 884. [Google Scholar] [CrossRef]
- Kim, M.S.; Chen, Y.; Mehl, P. Hyperspectral reflectance and fluorescence imaging system for food quality and safety. Trans. ASAE 2001, 44, 721. [Google Scholar]
- Jia, S.; Ji, Z.; Qian, Y.; Shen, L. Unsupervised band selection for hyperspectral imagery classification without manual band removal. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2012, 5, 531–543. [Google Scholar] [CrossRef]
- Zagoruyko, S.; Komodakis, N. Paying more attention to attention: Improving the performance of convolutional neural networks via attention transfer. arXiv 2016, arXiv:1612.03928. [Google Scholar]
- Li, Y.; Zhang, H.; Shen, Q. Spectral–spatial classification of hyperspectral imagery with 3D convolutional neural network. Remote Sens. 2017, 9, 67. [Google Scholar] [CrossRef]
- Hendrycks, D.; Gimpel, K. Gaussian error linear units (gelus). arXiv 2016, arXiv:1606.08415. [Google Scholar]









| Layer (Type) | Kernel | Stride | Padding | Input Size | Output Size |
|---|---|---|---|---|---|
| SeAM | 1 | - | |||
| SaAM | 1 | 3 | |||
| Rearrange | - | - | - | ||
| Conv3d-1 | 1 | same | |||
| BN1 + ReLU | - | - | - | ||
| Conv3d-2 | 1 | same | |||
| BN2 + ReLU | - | - | - | ||
| Linear Embedding | - | - | - | ||
| TE block | - | - | - | 64 | |
| Linear | - | - | - | 64 | 5 |
| Case | Component | Accuracy (%) | |||
|---|---|---|---|---|---|
| SeAM | SaAM | 3D Conv | TE | ||
| 1 | ✕ | ✕ | ✓ | ✕ | |
| 2 | ✕ | ✕ | ✕ | ✓ | |
| 3 | ✕ | ✕ | ✓ | ✓ | |
| 4 | ✕ | ✓ | ✓ | ✓ | |
| 5 | ✓ | ✕ | ✓ | ✓ | |
| 6 | ✓ | ✓ | ✕ | ✓ | |
| 7 | ✓ | ✓ | ✓ | ✕ | |
| 8 | ✓ | ✓ | ✓ | ✓ | |
| Model | Extraction Method | Number of Bands | Accuracy (%) | Precision (%) | Recall (%) | (%) |
|---|---|---|---|---|---|---|
| SVM | None | 288 | 92.62 | 92.61 | 92.71 | 92.66 |
| SPA | 46 | 85.63 | 85.60 | 85.48 | 85.54 | |
| UVE | 48 | 85.83 | 85.98 | 85.91 | 85.94 | |
| CARS | 31 | 91.26 | 91.54 | 91.20 | 91.37 | |
| LDA | None | 288 | 94.56 | 94.64 | 94.64 | 94.64 |
| SPA | 46 | 86.64 | 87.60 | 86.84 | 87.22 | |
| UVE | 48 | 89.78 | 89.90 | 89.90 | 89.90 | |
| CARS | 31 | 91.75 | 92.57 | 91.69 | 92.13 | |
| PLS–DA | None | 288 | 94.56 | 94.66 | 94.62 | 94.64 |
| SPA | 46 | 87.23 | 87.53 | 87.13 | 87.33 | |
| UVE | 48 | 90.37 | 90.90 | 90.22 | 90.56 | |
| CARS | 31 | 92.53 | 92.59 | 92.61 | 92.60 | |
| 1DCNN | None | 288 | 94.73 | 94.89 | 94.76 | 94.82 |
| SPA | 46 | 90.82 | 91.53 | 90.66 | 91.09 | |
| UVE | 48 | 92.92 | 93.25 | 92.74 | 92.99 | |
| CARS | 31 | 93.16 | 93.56 | 93.00 | 93.28 |
| Class | 2DCNN | 3DCNN | HybridSN | ResNet18 | SE–ResNet18 | SSA–3DTE |
|---|---|---|---|---|---|---|
| 1 | ||||||
| 2 | ||||||
| 3 | ||||||
| 4 | ||||||
| 5 | ||||||
| Accuracy (%) | ||||||
| Precision (%) | ||||||
| Recall (%) | ||||||
| (%) |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Xu, Z.; Hu, H.; Xu, H.; Zhao, Y.; Mao, X. Identification of Turtle-Shell Growth Year Using Hyperspectral Imaging Combined with an Enhanced Spatial–Spectral Attention 3DCNN and a Transformer. Molecules 2023, 28, 6427. https://doi.org/10.3390/molecules28176427
Wang T, Xu Z, Hu H, Xu H, Zhao Y, Mao X. Identification of Turtle-Shell Growth Year Using Hyperspectral Imaging Combined with an Enhanced Spatial–Spectral Attention 3DCNN and a Transformer. Molecules. 2023; 28(17):6427. https://doi.org/10.3390/molecules28176427
Chicago/Turabian StyleWang, Tingting, Zhenyu Xu, Huiqiang Hu, Huaxing Xu, Yuping Zhao, and Xiaobo Mao. 2023. "Identification of Turtle-Shell Growth Year Using Hyperspectral Imaging Combined with an Enhanced Spatial–Spectral Attention 3DCNN and a Transformer" Molecules 28, no. 17: 6427. https://doi.org/10.3390/molecules28176427
APA StyleWang, T., Xu, Z., Hu, H., Xu, H., Zhao, Y., & Mao, X. (2023). Identification of Turtle-Shell Growth Year Using Hyperspectral Imaging Combined with an Enhanced Spatial–Spectral Attention 3DCNN and a Transformer. Molecules, 28(17), 6427. https://doi.org/10.3390/molecules28176427