New Copper(II)-L-Dipeptide-Bathophenanthroline Complexes as Potential Anticancer Agents—Synthesis, Characterization and Cytotoxicity Studies—And Comparative DNA-Binding Study of Related Phen Complexes
Abstract
:1. Introduction
2. Results and Discussion
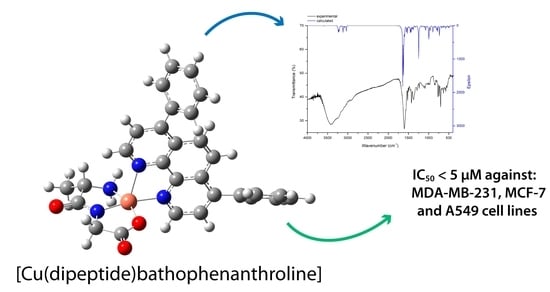
2.1. Geometry Optimization, IR Spectrum Calculation, and Interpretation
2.2. Solid-State Characterization: Infrared Spectra
2.3. Characterization in Solution UV–Visible Spectra
2.4. DNA binding
2.5. Cytotoxicity
3. Materials and Methods
3.1. Synthesis and Analytical Characterization
3.2. DFT Studies (Geometry Optimization and Infrared Spectra)
3.3. Spectroscopic Characterization
3.4. DNA Interaction
3.4.1. Determination of Kb via the UV Absorption Titration Experiments
3.4.2. Viscosity Studies
3.5. Cytotoxicity Studies
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Sung, H.; Ferlay, J.; Siegel, R.L.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries. CA Cancer J. Clin. 2021, 71, 209–249. [Google Scholar] [CrossRef]
- Chandrashekhar, M.; Nayak, V.L.; Ramakrishna, S.; Mallavadhani, U.V. Novel triazole hybrids of myrrhanone C, a natural polypodane triterpene: Synthesis, cytotoxic activity and cell based studies. Eur. J. Med. Chem. 2016, 114, 293–307. [Google Scholar] [CrossRef]
- Santini, C.; Pellei, M.; Gandin, V.; Porchia, M.; Tisato, F.; Marzano, C. Advances in Copper Complexes as Anticancer Agents. Chem. Rev. 2014, 114, 815–862. [Google Scholar] [CrossRef] [PubMed]
- Kellett, A.; Molphy, Z.; McKee, V.; Slator, C. Recent Advances in Anticancer Copper Compounds. In Metal-Based Anticancer Agents; RSC Publishing: Cambridge, UK, 2019; pp. 91–119. [Google Scholar]
- Van Rijt, S.H.; Sadler, P.J. Current applications and future potential for bioinorganic chemistry in the development of anticancer drugs. Drug Discov. Today 2009, 14, 1089–1097. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Casini, A.; Vessières, A.; Meier-Menches, S.M. Metal-Based Anticancer Agents; Royal Society of Chemistry: London, UK, 2019; Volume 14. [Google Scholar]
- Figueroa-DePaz, Y.; Pérez-Villanueva, J.; Soria-Arteche, O.; Martínez-Otero, D.; Gómez-Vidales, V.; Ortiz-Frade, L.; Ruiz-Azuara, L. Casiopeinas of Third Generations: Synthesis, Characterization, Cytotoxic Activity and StructureߞActivity Relationships of Mixed Chelate Compounds with Bioactive Secondary Ligands. Molecules 2022, 27, 3504. [Google Scholar] [PubMed]
- Aguilar-Jimenez, Z.; Gonzalez-Ballesteros, M.; Davila-Manzanilla, S.G.; Espinoza-Guillen, A.; Ruiz-Azuara, L. Development and In Vitro and In Vivo Evaluation of an Antineoplastic Copper(II) Compound (Casiopeina III-ia) Loaded in Nonionic Vesicles Using Quality by Design. Int. J. Mol. Sci. 2022, 23, 12756. [Google Scholar] [CrossRef] [PubMed]
- Marzano, C.; Gandin, V.; Pellei, M.; Colavito, D.; Papini, G.; Lobbia, G.G.; Del Giudice, E.; Porchia, M.; Tisato, F.; Santini, C. In vitro antitumor activity of the water soluble copper(I) complexes bearing the tris(hydroxymethyl)phosphine ligand. J. Med. Chem. 2008, 51, 798–808. [Google Scholar] [CrossRef]
- Gandin, V.; Ceresa, C.; Esposito, G.; Indraccolo, S.; Porchia, M.; Tisato, F.; Santini, C.; Pellei, M.; Marzano, C. Therapeutic potential of the phosphino Cu(I) complex (HydroCuP) in the treatment of solid tumors. Sci. Rep. 2017, 7, 13936. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ceresa, C.; Nicolini, G.; Semperboni, S.; Gandin, V.; Monfrini, M.; Avezza, F.; Alberti, P.; Bravin, A.; Pellei, M.; Santini, C.; et al. Evaluation of the Profile and Mechanism of Neurotoxicity of Water-Soluble [Cu(P)(4)]PF(6) and [Au(P)(4)]PF(6) (P = thp or PTA) Anticancer Complexes. Neurotox. Res. 2018, 34, 93–108. [Google Scholar] [CrossRef]
- Northcote-Smith, J.; Kaur, P.; Suntharalingam, K. A Cancer Stem Cell Potent Copper(II) Complex with a S, N, S-Schiff base Ligand and Bathophenanthroline. Eur. J. Inorg. Chem. 2021, 2021, 1770–1775. [Google Scholar] [CrossRef]
- Kaur, P.; Johnson, A.; Northcote-Smith, J.; Lu, C.; Suntharalingam, K. Immunogenic Cell Death of Breast Cancer Stem Cells Induced by an Endoplasmic Reticulum-Targeting Copper(II) Complex. ChemBioChem 2020, 21, 3618–3624. [Google Scholar] [CrossRef]
- Mejía, C.; Ortega-Rosales, S.; Ruiz-Azuara, L. Mechanism of Action of Anticancer Metallodrugs. In Biomedical Applications of Metals; Springer: Cham, Switzerland, 2018; pp. 213–234. [Google Scholar]
- Masuri, S.; Vaňhara, P.; Cabiddu, M.G.; Moráň, L.; Havel, J.; Cadoni, E.; Pivetta, T. Copper(II) Phenanthroline-Based Complexes as Potential AntiCancer Drugs: A Walkthrough on the Mechanisms of Action. Molecules 2022, 27, 49. [Google Scholar] [CrossRef]
- Molinaro, C.; Martoriati, A.; Pelinski, L.; Cailliau, K. Copper Complexes as Anticancer Agents Targeting Topoisomerases I and II. Cancers 2020, 12, 2863. [Google Scholar] [CrossRef] [PubMed]
- Tsvetkov, P.; Coy, S.; Petrova, B.; Dreishpoon, M.; Verma, A.; Abdusamad, M.; Rossen, J.; Joesch-Cohen, L.; Humeidi, R.; Spangler, R.D.; et al. Copper induces cell death by targeting lipoylated TCA cycle proteins. Science 2022, 375, 1254–1261. [Google Scholar] [CrossRef]
- Iglesias, S.; Alvarez, N.; Torre, M.H.; Kremer, E.; Ellena, J.; Ribeiro, R.R.; Barroso, R.P.; Costa-Filho, A.J.; Kramer, M.G.; Facchin, G. Synthesis, structural characterization and cytotoxic activity of ternary copper (II)–dipeptide–phenanthroline complexes. A step towards the development of new copper compounds for the treatment of cancer. J. Inorg. Biochem. 2014, 139, 117–123. [Google Scholar] [CrossRef]
- Iglesias, S.; Alvarez, N.; Kramer, G.; Torre, M.H.; Kremer, E.; Ellena, J.; Costa-Filho, A.J.; Facchin, G. Structural Characterization and Cytotoxic Activity of Heteroleptic Copper (II) Complexes with L-Dipeptides and 5-NO2-Phenanthroline. Crystal Structure of [Cu (Phe-Ala)(5-NO2-Phen)]. 4H2O. Struct. Chem. Crystallogr. Commun. 2015, 1, 7. [Google Scholar]
- Alvarez, N.; Viña, D.; Leite, C.M.; Mendes, L.F.; Batista, A.A.; Ellena, J.; Costa-Filho, A.J.; Facchin, G. Synthesis and structural characterization of a series of ternary copper (II)-L-dipeptide-neocuproine complexes. Study of their cytotoxicity against cancer cells including MDA-MB-231, triple negative breast cancer cells. J. Inorg. Biochem. 2020, 203, 110930. [Google Scholar] [CrossRef] [PubMed]
- Veiga, N.; Alvarez, N.; Castellano, E.E.; Ellena, J.; Facchin, G.; Torre, M.H. Comparative Study of Antioxidant and Pro-Oxidant Properties of Homoleptic and Heteroleptic Copper Complexes with Amino Acids, Dipeptides and 1,10-Phenanthroline: The Quest for Antitumor Compounds. Molecules 2021, 26, 6520. [Google Scholar] [CrossRef] [PubMed]
- Alvarez, N.; Leite, C.M.; Napoleone, A.; Mendes, L.F.S.; Fernandez, C.Y.; Ribeiro, R.R.; Ellena, J.; Batista, A.A.; Costa-Filho, A.J.; Facchin, G. Tetramethyl-phenanthroline copper complexes in the development of drugs to treat cancer: Synthesis, characterization and cytotoxicity studies of a series of copper(II)-L-dipeptide-3,4,7,8-tetramethyl-phenanthroline complexes. J. Biol. Inorg. Chem. 2022, 27, 431–441. [Google Scholar] [CrossRef]
- Facchin, G.; Veiga, N.; Kramer, M.G.; Batista, A.A.; Várnagy, K.; Farkas, E.; Moreno, V.; Torre, M.H. Experimental and theoretical studies of copper complexes with isomeric dipeptides as novel candidates against breast cancer. J. Inorg. Biochem. 2016, 162, 52–61. [Google Scholar] [CrossRef]
- Facchin, G.; Torre, M.H.; Kremer, E.; Piro, O.E.; Castellano, E.E.; Baran, E.J. Structural and spectroscopic characterization of two new Cu (II)-dipeptide complexes. Z. Fur Nat. B 2000, 55, 1157–1162. [Google Scholar] [CrossRef]
- Facchin, G.; Torre, M.H.; Kremer, E.; Piro, O.E.; Castellano, E.E.; Baran, E.J. Synthesis and characterization of three new Cu(II)-dipeptide complexes. J. Inorg. Biochem. 2002, 89, 174–180. [Google Scholar] [CrossRef] [PubMed]
- Facchin, G.; Torre, M.; Kremer, E.; Baran, E.; Mombrú, A.; Pardo, H.; Araujo, M.; Batista, A.; Costa-Filho, A. Cu (II) complexation with His–Gly and His–Ala. X-ray structure of [Cu (his–gly)2(H2O)2]· 6H2O. Inorg. Chim. Acta 2003, 355, 408–413. [Google Scholar] [CrossRef]
- Sigel, H.; Martin, R.B. Coordinating properties of the amide bond. Stability and structure of metal ion complexes of peptides and related ligands. Chem. Rev. 1982, 82, 385–426. [Google Scholar] [CrossRef]
- Prenesti, E.; Daniele, P.; Prencipe, M.; Ostacoli, G. Spectrum–structure correlation for visible absorption spectra of copper (II) complexes in aqueous solution. Polyhedron 1999, 18, 3233–3241. [Google Scholar] [CrossRef]
- Prenesti, E.; Daniele, P.G.; Berto, S.; Toso, S. Spectrum–structure correlation for visible absorption spectra of copper (II) complexes showing axial co-ordination in aqueous solution. Polyhedron 2006, 25, 2815–2823. [Google Scholar] [CrossRef]
- Rehman, S.U.; Sarwar, T.; Husain, M.A.; Ishqi, H.M.; Tabish, M. Studying non-covalent drug-DNA interactions. Arch. Biochem. Biophys. 2015, 576, 49–60. [Google Scholar] [CrossRef] [PubMed]
- Suh, D.; Chaires, J.B. Criteria for the mode of binding of DNA binding agents. Bioorg. Med. Chem. 1995, 3, 723–728. [Google Scholar] [CrossRef]
- Satyanarayana, S.; Dabrowiak, J.C.; Chaires, J.B. Neither delta- nor lambda-tris(phenanthroline)ruthenium(II) binds to DNA by classical intercalation. Biochemistry 1992, 31, 9319–9324. [Google Scholar] [CrossRef] [PubMed]
- Kapicak, L.; Gabbay, E.J. Effect of aromatic cations on the tertiary structure of deoxyribonucleic acid. J. Am. Chem. Soc. 1975, 97, 403–408. [Google Scholar] [CrossRef]
- Gratal, P.; Arias-Perez, M.S.; Gude, L. 1H-imidazo[4,5-f][1,10]phenanthroline carbohydrate conjugates: Synthesis, DNA interactions and cytotoxic activity. Bioorg. Chem. 2022, 125, 105851. [Google Scholar] [CrossRef]
- Chikira, M.; Ng, C.H.; Palaniandavar, M. Interaction of DNA with Simple and Mixed Ligand Copper (II) Complexes of 1, 10-Phenanthrolines as Studied by DNA-Fiber EPR Spectroscopy. Int. J. Mol. Sci. 2015, 16, 22754–22780. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Figueroa-DePaz, Y.; Resendiz-Acevedo, K.; Davila-Manzanilla, S.G.; Garcia-Ramos, J.C.; Ortiz-Frade, L.; Serment-Guerrero, J.; Ruiz-Azuara, L. DNA, a target of mixed chelate copper(II) compounds (Casiopeinas(R)) studied by electrophoresis, UV-vis and circular dichroism techniques. J. Inorg. Biochem. 2022, 231, 111772. [Google Scholar] [CrossRef]
- Mahadevan, S.; Palaniandavar, M. Spectral and Electrochemical Behavior of Copper(II)-Phenanthrolines Bound to Calf Thymus DNA. [(5,6-dimethyl-OP)(2)Cu](2+) (5,6-dimethyl-OP = 5,6-Dimethyl-1,10-phenanthroline) Induces a Conformational Transition from B to Z DNA. Inorg. Chem. 1998, 37, 3927–3934. [Google Scholar] [CrossRef]
- Facchin, G.; Kremer, E.; Baran, E.J.; Castellano, E.E.; Piro, O.E.; Ellena, J.; Costa-Filho, A.J.; Torre, M.H. Structural characterization of a series of new Cu-dipeptide complexes in solid state and in solution. Polyhedron 2006, 25, 2597–2604. [Google Scholar] [CrossRef]
- Dennington, R.; Keith, T.; Millam, J. GaussView, 5.0; Semichem Inc.: Shawnee Mission, KS, USA, 2009. [Google Scholar]
- Lewars, E.G. Introduction to Quantum Mechanics in Computational Chemistry. In Computational Chemistry: Introduction to the Theory and Applications of Molecular and Quantum Mechanics; Springer International Publishing: Cham, Switzerland, 2016; pp. 101–191. [Google Scholar]
- Lee, C.; Yang, W.; Parr, R.G. Development of the Colle-Salvetti correlation-energy formula into a functional of the electron density. Phys. Rev. B Condens. Matter 1988, 37, 785–789. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wadt, W.R.; Hay, P.J. Ab initio effective core potentials for molecular calculations. Potentials for main group elements Na to Bi. J. Chem. Phys. 1985, 82, 284–298. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for K to Au including the outermost core orbitals. J. Chem. Phys. 1985, 82, 299–310. [Google Scholar] [CrossRef]
- Hay, P.J.; Wadt, W.R. Ab initio effective core potentials for molecular calculations. Potentials for the transition metal atoms Sc to Hg. J. Chem. Phys. 1985, 82, 270–283. [Google Scholar] [CrossRef]
- Frisch, M.J.; Trucks, G.W.; Schlegel, H.B.; Scuseria, G.E.; Robb, M.A.; Cheeseman, J.R.; Scalmani, G.; Barone, V.; Mennucci, B.; Petersson, G.A.; et al. Gaussian 09; Gaussian, Inc.: Wallingford, CT, USA, 2009. [Google Scholar]
- Jamróz, M.H. Vibrational Energy Distribution Analysis (VEDA): Scopes and limitations. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2013, 114, 220–230. [Google Scholar] [CrossRef]
- Benesi, H.A.; Hildebrand, J.H. A Spectrophotometric Investigation of the Interaction of Iodine with Aromatic Hydrocarbons. J. Am. Chem. Soc. 2002, 71, 2703–2707. [Google Scholar] [CrossRef]
- Scruggs, R.L.; Ross, P.D. Viscosity study of DNA. Biopolymers 1964, 2, 593–609. [Google Scholar] [CrossRef]




| Experimental | Calculated | PED% | Assignment |
|---|---|---|---|
| 3412 | 3583 | 99 | νas(N-H) |
| 3477 | 99 | νs(N-H) | |
| 3240 | 3252 | 98 | νs, batho(C-H) |
| 3244 | 94 | ||
| 3238 | 80 | ||
| 3236 | 82 | ||
| 3235 | 82 | ||
| 3231 | 94 | νas, batho(C-H) | |
| 3221 | 81 | ||
| 3220 | 61 | ||
| 3220 | 85 | ||
| 3141 | 3212 | 79 | νas,batho(C-H) |
| 3211 | 93 | ||
| 3210 | 82 | ||
| 3200 | 85 | ||
| 3199 | 83 | ||
| 3195 | 84 | ||
| 3194 | 88 | ||
| 3163 | 84 | νas, dipeptide(C-H) | |
| 3117 | 93 | ||
| 2917 | 3083 | 99 | νas, dipeptide(C-H) |
| 3068 | 94 | ||
| 3038 | 99 | νs, dipeptide(C-H) | |
| 3033 | 84 | ||
| 1597 | 1683 | 77 | δ(H-N-H) dipeptide |
| 1662 | 44 | ν(C-C) batho | |
| 1653 | 52 | ||
| 1652 | 53 | ||
| 1642 | 80 | νas(COO) dipeptide | |
| 1564 | 1621 | 81 | ν(N-C) + ν(O-C) dipeptide |
| 1593 | 32 | ν(N-C) batho | |
| 1521 | 1526 | 49 | δ(H-C-C) batho + dipeptide |
| 1526 | 64 | δ(H-C-H) dipeptide | |
| 1521 | 51 | δ(H-C-C) batho | |
| 1492 | 1508 | 72 | δ(H-C-H) dipeptide |
| 1499 | 68 | ||
| 1499 | 29 | τ(H-C-C-O) dipeptide | |
| 1475 | 49 | δ(H-C-C) batho | |
| 1472 | 39 | ||
| 1427 | 1433 | 87 | δ(H-C-H) methyl in dipeptide |
| 1404 | 46 | ν(N-C) + ν(O-C) + ν(C-C) dipeptide | |
| 1374 | 1382 | 40 | τ(H-C-C-N) dipeptide |
| 1379 | 46 | δ(H-C-C) batho | |
| 1378 | 29 | ν(C-C) batho | |
| 1378 | 42 | δ(H-C-C) batho | |
| 1352 | 25 | ν(C-C) batho | |
| 1288 | 1335 | 25 | δ(H-C-H) dipeptide |
| 1335 | 50 | τ(H-C-C-O) dipeptide | |
| 1323 | 55 | δ(H-C-C) dipeptide | |
| 1232 | 1248 | 58 | νs(COO) + ν(C-C) dipeptide |
| 1228 | 67 | δ(H-C-C) batho | |
| 1227 | 65 | ||
| 1183 | 1215 | 27 | δ(H-C-C) batho |
| 1212 | 49 | δ(H-C-C) dipeptide | |
| 1212 | 31 | τ(H-C-C-O) dipeptide | |
| 1209 | 78 | δ(H-C-C) batho | |
| 1209 | 78 | δ(H-C-C) batho | |
| 1157 | 1130 | 55 | ν(N-C) + ν(C-C) dipeptide |
| 1090 | 1073 | 49 | ν(N-C) + ν(C-C) dipeptide |
| 1022 | 1047 | 28 | τoop(H-C-C-C) batho |
| 1045 | 62 | ||
| 1042 | 47 | ||
| 1025 | 76 | ||
| 1024 | 27 | δ(H-N-C) batho | |
| 1024 | 34 | τoop(H-C-C-N) batho | |
| 1023 | 73 | τoop(H-C-C-C) batho | |
| 999 | 1012 | 43 | τoop(H-C-C-C) batho |
| 1010 | 26 | δ(H-C-C) dipeptide | |
| 1006 | 73 | τoop(H-C-C-C) batho | |
| 997 | 53 | τ(H-N-C-C) dipeptide + batho | |
| 972 | 970 | 75 | τoop(H-C-C-C) batho |
| 969 | 69 | ||
| 910 | 40 | δ(C-C-C) + δ(C-C-N) batho | |
| 858 | 903 | 68 | τoop(H-C-C-C) batho |
| 897 | 84 | ||
| 895 | 50 | τoop(H-C-C-N) batho | |
| 893 | 25 | ||
| 891 | 65 | ||
| 890 | 64 | ν(N-C) + ν(C-C) + ν(O-C) dipeptide | |
| 882 | 66 | τoop(H-C-C-C) batho | |
| 843 | 851 | 45 | ν(N-C) + ν(C-C) + ν(O-C) dipeptide |
| 810 | 799 | 38 | τoop(H-C-C-C) batho |
| 768 | 743 | 48 | τ(O-C-N-C) dipeptide |
| 740 | 732 | 48 | τoop(H-C-C-C) batho |
| 732 | 29 | τ(C-C-C-C) batho | |
| 731 | 44 | τoop(H-C-C-C) batho | |
| 731 | 28 | τ(C-C-C-C) batho | |
| 704 | 682 | 61 | δ(C-C-O) dipeptide |
| 666 | 640 | 39 | δ(C-C-C) batho |
| 630 | 632 | 28 | δ(C-C-C) batho |
| 598 | 581 | 47 | τ(H-N-C-C) dipeptide |
| 575 | 564 | 64 | τ(O-C-O-C) dipeptide |
| 548 | 543 | 26 | δ(C-C-N) dipeptide |
| 521 | 507 | 52 | δ(C-C-N) + δ(C-C-O) + δ(O-C-O) dipeptide |
| 438 | 428 | 37 | τ(C-N-C-C) dipeptide |
| 417 | 421 | 74 | τ(C-C-C-C) batho |
| 420 | 72 | τ(C-C-C-C) batho |
| Compound | νs + νas(N-H) * | ν(C=O) * + ν(C-N) ** + ν as (COO) * + ν(C-C) ** | ν(N-C) * + ν(C=O) * + ν(C-C) * | νs(COO) * + ν(C-C) * | ρ(C-H) ** | δ(C-C-O)* | δ(C-C-N) * + δ(C-C-O) * + δ(O-C-O) * |
|---|---|---|---|---|---|---|---|
| 1 | 3415sh | 1588s, 1516w | 1420m | 1239w | 1040w | 704s | 536w |
| 2 | 3401sh | 1594s, 1516w | 1417m | 1226w | 1040w | 704s | 542w |
| 3 | 3412sh | 1597s, 1521w | 1427m | 1232w | 1094w | 704s | 575w |
| 4 | 3415sh | 1594s, 1516w | 1413m | 1226w | 1084w | 704s | 549w |
| 5 | 3401sh | 1601s, 1523w | 1413m | 1233w | 1065w | 704s | 542w |
| 6 | 3415sh | 1601s, 1516w | 1420m | 1239w | 1064w | 704s | 549w |
| 7 | 3408sh | 1594s, 1516w | 1427m | 1233m | 1065w | 704s | 536w |
| 8 | 3401sh | 1594s, 1523w | 1413m | 1233m | 1072w | 704s | 555w |
| Compound | λmax (nm) in DMSO/in Water:DMSO 50:50 * | ɛ (DMSO) |
|---|---|---|
| 1 | 608 | 178 |
| 2 | 610 | 147 |
| 3 | 615/630 | 132 |
| 4 | 611 | 92 |
| 5 | 610/620 | 133 |
| 6 | 611 | 142 |
| 7 | 610 | 106 |
| 8 | 608 | 121 |
| Compound | MDA-MB-231 | MCF-7 | MCF-10A | A549 | MRC-5 | |
|---|---|---|---|---|---|---|
| 1 | [Cu(gly-val)(batho)] | 0.41 ± 0.03 | 0.40 ± 0.01 | 4.95 ± 0.94 | 1.20 ± 0.36 | 0.60 ± 0.12 |
| 2 | [Cu(gly-phe)(batho)] | 1.16 ± 0.22 | 1.40 ± 0.20 | 5.28 ± 0.52 | 1.77 ± 0.22 | 1.72 ± 0.09 |
| 3 | [Cu(ala-gly)(batho)] | 5.30 ± 0.81 | 1.34 ± 0.80 | 6.67 ± 1.89 | 2.18 ± 0.44 | 0.15 ± 0.03 |
| 4 | [Cu(ala-ala)(batho)] | 0.47 ± 0.07 | 1.45 ± 0.43 | 3.60 ± 0.31 | 0.90 ± 0.07 | 0.61 ± 0.17 |
| 5 | [Cu(ala-phe)(batho)] | 0.97 ± 0.20 | 1.49 ± 0.46 | 3.86 ± 0.98 | 0.66 ± 0.20 | 0.31 ± 0.05 |
| 6 | [Cu(phe-ala)(batho)] | 0.79 ± 0.13 | 0.53 ± 0.09 | 1.78 ± 0.41 | 0.33 ± 0.13 | 0.25 ± 0.02 |
| 7 | [Cu(phe-val)(batho)] | 0.65 ± 0.06 | 1.54 ± 0.96 | 3.75 ± 0.78 | 1.47 ± 0.16 | 0.35 ± 0.05 |
| 8 | [Cu(phe-phe)(batho)] | 1.06 ± 0.44 | 3.34 ± 2.38 | 4.28 ± 0.75 | 1.59 ± 0.18 | 0.99 ± 0.21 |
| 0 | [CuCl2(batho)] | 0.47 ± 0.07 | 2.75 ± 0.84 | 2.75 ± 0.60 | 0.87 ± 0.11 | 0.85 ± 0.15 |
| Cisplatin | 12.43 ± 0.20 | 8.91 ± 2.60 | 23.90 ± 0.70 | 14.40 ± 1.40 | 29.09 ± 0.78 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Fernández, C.Y.; Alvarez, N.; Rocha, A.; Ellena, J.; Costa-Filho, A.J.; Batista, A.A.; Facchin, G. New Copper(II)-L-Dipeptide-Bathophenanthroline Complexes as Potential Anticancer Agents—Synthesis, Characterization and Cytotoxicity Studies—And Comparative DNA-Binding Study of Related Phen Complexes. Molecules 2023, 28, 896. https://doi.org/10.3390/molecules28020896
Fernández CY, Alvarez N, Rocha A, Ellena J, Costa-Filho AJ, Batista AA, Facchin G. New Copper(II)-L-Dipeptide-Bathophenanthroline Complexes as Potential Anticancer Agents—Synthesis, Characterization and Cytotoxicity Studies—And Comparative DNA-Binding Study of Related Phen Complexes. Molecules. 2023; 28(2):896. https://doi.org/10.3390/molecules28020896
Chicago/Turabian StyleFernández, Carlos Y., Natalia Alvarez, Analu Rocha, Javier Ellena, Antonio J. Costa-Filho, Alzir A. Batista, and Gianella Facchin. 2023. "New Copper(II)-L-Dipeptide-Bathophenanthroline Complexes as Potential Anticancer Agents—Synthesis, Characterization and Cytotoxicity Studies—And Comparative DNA-Binding Study of Related Phen Complexes" Molecules 28, no. 2: 896. https://doi.org/10.3390/molecules28020896
APA StyleFernández, C. Y., Alvarez, N., Rocha, A., Ellena, J., Costa-Filho, A. J., Batista, A. A., & Facchin, G. (2023). New Copper(II)-L-Dipeptide-Bathophenanthroline Complexes as Potential Anticancer Agents—Synthesis, Characterization and Cytotoxicity Studies—And Comparative DNA-Binding Study of Related Phen Complexes. Molecules, 28(2), 896. https://doi.org/10.3390/molecules28020896