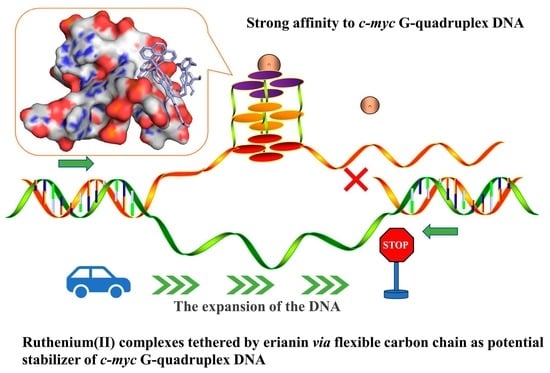
Ruthenium(II) Complexes Coupled by Erianin via a Flexible Carbon Chain as a Potential Stabilizer of c-myc G-Quadruplex DNA
Abstract
:1. Introduction
2. Results and Discussion
2.1. Synthesis and Characterization
2.2. DNA Binding Behaviors
2.2.1. Electronic Spectra Titration Experiments
2.2.2. Fluorescence Response towards c-myc G-Quadruplex DNA
2.2.3. Circular Dichroism (CD) Spectroscopy
2.2.4. Melting FRET and Competitive FRET Assays
2.2.5. PCR-Stop Assay
2.2.6. Molecular Docking
3. Materials and Methods
3.1. Chemicals
3.2. Instruments
3.3. Synthesis and Characterization of [Ru(phen)2(L1(CH2)4Br)] (ClO4)2 (1b)
3.4. Synthesis and Characterization of [Ru(phen)2(L2(CH2)4Br)] (ClO4)2 (2b)
3.5. Synthesis and Characterization of [Ru(phen)2(L1(CH2)4eria)] (ClO4)2 (1)
3.6. Synthesis and Characterization of [Ru(phen)2(L2(CH2)4eria)] (ClO4)2 (2)
3.7. Electronic Spectra Titration Experiments
3.8. Fluorescence Spectra
3.9. Fluorescence Quantum Yields
3.10. Circular Dichroism (CD) Spectroscopy
3.11. FRET Melting and Competitive FRET Assays
3.12. Polymerase Chain Reaction (PCR-Stop) Assay
3.13. Molecular Docking
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Sample Availability
References
- Wang, Y.; Yang, J.; Wild, A.T.; Wu, W.H.; Shah, R.; Danussi, C.; Riggins, G.J.; Kannan, K.; Sulman, E.P.; Chan, T.A.; et al. G-quadruplex DNA drives genomic instability and represents a targetable molecular abnormality in ATRX-deficient malignant glioma. Nat. Commun. 2019, 10, 943. [Google Scholar] [CrossRef]
- Tauchi, T.; Shin-Ya, K.; Sashida, G.; Sumi, M.; Nakajima, A.; Shimamoto, T.; Ohyashiki, J.H.; Ohyashiki, K. Activity of a novel G-quadruplex-interactive telomerase inhibitor, telomestatin (SOT-095), against human leukemia cells: Involvement of ATM-dependent DNA damage response pathways. Oncogene 2003, 22, 5338–5347. [Google Scholar] [CrossRef] [PubMed]
- Wheelhouse, R.T.; Sun, D.; Han, H.; Han, F.X.; Hurley, L.H. Cationic Porphyrins as Telomerase Inhibitors: The Interaction of Tetra-(N-methyl-4-pyridyl)porphine with Quadruplex DNA. J. Am. Chem. Soc. 1998, 120, 3261–3262. [Google Scholar] [CrossRef]
- Phatak, P.; Cookson, J.C.; Dai, F.; Smith, V.; Gartenhaus, R.B.; Stevens, M.F.; Burger, A.M. Telomere uncapping by the G-quadruplex ligand RHPS4 inhibits clonogenic tumour cell growth in vitro and in vivo consistent with a cancer stem cell targeting mechanism. Br. J. Cancer 2007, 96, 1223–1233. [Google Scholar] [CrossRef]
- Siddiqui-Jain, A.; Grand, C.L.; Bearss, D.J.; Hurley, L.H. Direct evidence for a G-quadruplex in a promoter region and its targeting with a small molecule to repress c-MYC transcription. Proc. Natl. Acad. Sci. USA 2002, 99, 11593–11598. [Google Scholar] [CrossRef] [PubMed]
- Varshney, D.; Spiegel, J.; Zyner, K.; Tannahill, D.; Balasubramanian, S. The regulation and functions of DNA and RNA G-quadruplexes. Nat. Rev. Mol. Cell Biol. 2020, 21, 459–474. [Google Scholar] [CrossRef]
- Chen, B.-J.; Wu, Y.-L.; Tanaka, Y.; Zhang, W. Small Molecules Targeting c-Myc Oncogene: Promising Anti-Cancer Therapeutics. Int. J. Biol. Sci. 2014, 10, 1084–1096. [Google Scholar] [CrossRef] [PubMed]
- Mathad, R.I.; Hatzakis, E.; Dai, J.; Yang, D. c-MYC promoter G-quadruplex formed at the 5′-end of NHE III1 element: Insights into biological relevance and parallel-stranded G-quadruplex stability. Nucleic Acids Res. 2011, 39, 9023–9033. [Google Scholar] [CrossRef]
- McKeown, M.R.; Bradner, J.E. Therapeutic strategies to inhibit MYC. Cold Spring Harb. Perspect. Med. 2014, 4, a014266. [Google Scholar] [CrossRef]
- Rangan, A.; Fedoroff, O.Y.; Hurley, L.H. Induction of duplex to G-quadruplex transition in the c-myc promoter region by a small molecule. J. Biol. Chem. 2001, 276, 4640–4646. [Google Scholar] [CrossRef] [Green Version]
- Burger, A.M.; Dai, F.; Schultes, C.M.; Reszka, A.P.; Moore, M.J.; Double, J.A.; Neidle, S. The G-quadruplex-interactive molecule BRACO-19 inhibits tumor growth, consistent with telomere targeting and interference with telomerase function. Cancer Res. 2005, 65, 1489–1496. [Google Scholar] [CrossRef]
- Seenisamy, J.; Bashyam, S.; Gokhale, V.; Vankayalapati, H.; Sun, D.; Siddiqui-Jain, A.; Streiner, N.; Shin-Ya, K.; White, E.; Wilson, W.D.; et al. Design and synthesis of an expanded porphyrin that has selectivity for the c-MYC G-quadruplex structure. J. Am. Chem. Soc. 2005, 127, 2944–2959. [Google Scholar] [CrossRef]
- Gunaratnam, M.; de la Fuente, M.; Hampel, S.M.; Todd, A.K.; Reszka, A.P.; Schätzlein, A.; Neidle, S. Targeting pancreatic cancer with a G-quadruplex ligand. Bioorg. Med. Chem. 2011, 19, 7151–7157. [Google Scholar] [CrossRef] [PubMed]
- Drygin, D.; Siddiqui-Jain, A.; O′Brien, S.; Schwaebe, M.; Lin, A.; Bliesath, J.; Ho, C.B.; Proffitt, C.; Trent, K.; Whitten, J.P.; et al. Anticancer activity of CX-3543: A direct inhibitor of rRNA biogenesis. Cancer Res. 2009, 69, 7653–7661. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Liu, Y.; Liu, D.; Zhang, R.; Yang, X.; Liu, J. Stabilization of G-quadruplex DNA, inhibition of telomerase activity and live cell imaging studies of chiral ruthenium(II) complexes. Chemistry 2012, 18, 4285–4295. [Google Scholar] [CrossRef]
- Chen, X.; Wu, J.H.; Lai, Y.W.; Zhao, R.; Chao, H.; Ji, L.N. Targeting telomeric G-quadruplexes with the ruthenium(II) complexes [Ru(bpy)(2)(ptpn)](2+) and [Ru(phen)(2)(ptpn)](2+). Dalton Trans. 2013, 42, 4386–4397. [Google Scholar] [CrossRef] [PubMed]
- Sun, D.; Zhang, R.; Yuan, F.; Liu, D.; Zhou, Y.; Liu, J. Studies on characterization, telomerase inhibitory properties and G-quadruplex binding of η6-arene ruthenium complexes with 1,10-phenanthroline-derived ligands. Dalton Trans. 2012, 41, 1734–1741. [Google Scholar] [CrossRef]
- Shi, S.; Liu, J.; Yao, T.; Geng, X.; Jiang, L.; Yang, Q.; Cheng, L.; Ji, L. Promoting the formation and stabilization of G-quadruplex by dinuclear RuII complex Ru2(obip)L4. Inorg. Chem. 2008, 47, 2910–2912. [Google Scholar] [CrossRef]
- Zhao, H.; Xu, X.; Wang, S.; Mi, Y.; Zheng, Z.; Zhao, X. A dinuclear ruthenium(II) complex as an inducer and potential luminescent switch-on probe for G-quadruplex DNA. Transit. Met. Chem. 2018, 43, 539–548. [Google Scholar] [CrossRef]
- McQuaid, K.T.; Takahashi, S.; Baumgaertner, L.; Cardin, D.J.; Paterson, N.G.; Hall, J.P.; Sugimoto, N.; Cardin, C.J. Ruthenium Polypyridyl Complex Bound to a Unimolecular Chair-Form G-Quadruplex. J. Am. Chem. Soc. 2022, 144, 5956–5964. [Google Scholar] [CrossRef]
- Wang, C.; Yu, Q.; Yang, L.; Liu, Y.; Sun, D.; Huang, Y.; Zhou, Y.; Zhang, Q.; Liu, J. Ruthenium (II) polypyridyl complexes stabilize the bcl-2 promoter quadruplex and induce apoptosis of Hela tumor cells. Biometals 2013, 26, 387–402. [Google Scholar] [CrossRef]
- Zhang, Z.; Wu, Q.; Wu, X.H.; Sun, F.Y.; Chen, L.M.; Chen, J.C.; Yang, S.L.; Mei, W.J. Ruthenium(II) complexes as apoptosis inducers by stabilizing c-myc G-quadruplex DNA. Eur. J. Med. Chem. 2014, 80, 316–324. [Google Scholar] [CrossRef]
- Li, L.; Liu, H.-M.; Liu, X.-K.; Liao, S.-Y.; Lan, Y.-T.; Wu, Q.; Wang, X.-C.; Wang, Q.; Zhang, S.-Y.; Mei, W.-J. A Ruthenium(ii) complex as a potential luminescent switch-on probe for G-quadruplex DNA. RSC Adv. 2017, 7, 23727–23734. [Google Scholar] [CrossRef]
- Wu, Q.; Chen, T.; Zhang, Z.; Liao, S.; Wu, X.; Wu, J.; Mei, W.; Chen, Y.; Wu, W.; Zeng, L.; et al. Microwave-assisted synthesis of arene ruthenium(II) complexes [(η⁶-RC₆H₅)Ru(m-MOPIP)Cl]Cl (R = -H and -CH₃) as groove binder to c-myc G4 DNA. Dalton Trans. 2014, 43, 9216–9225. [Google Scholar] [CrossRef]
- Wu, Q.; Song, Y.; Liu, R.; Wang, R.; Mei, W.; Chen, W.; Yang, H.; Wang, X. Synthesis, docking studies and antitumor activity of phenanthroimidazole derivatives as promising c-myc G-quadruplex DNA stabilizers. Bioorg. Chem. 2020, 102, 104074. [Google Scholar] [CrossRef]
- Zhang, S.; Wu, Q.; Zhang, H.; Wang, Q.; Wang, X.; Mei, W.; Wu, X.; Zheng, W. Microwave-assisted synthesis of ruthenium(II) complexes with alkynes as potential inhibitor by selectively recognizing c-myc G-quadruplex DNA. J. Inorg. Biochem. 2017, 176, 113–122. [Google Scholar] [CrossRef] [PubMed]
- Yuan, C.; Wang, Z.; Wang, Z.; Liu, W.; Li, G.; Meng, J.; Wu, R.; Wu, Q.; Wang, J.; Mei, W. Novel Chiral Ru(II) Complexes as Potential c-myc G-quadruplex DNA Stabilizers Inducing DNA Damage to Suppress Triple-Negative Breast Cancer Progression. Int. J. Mol. Sci. 2022, 24, 203. [Google Scholar] [CrossRef] [PubMed]
- Qian, J.; Liu, R.; Liu, N.; Yuan, C.; Wu, Q.; Chen, Y.; Tan, W.; Mei, W. Arene Ru(II) Complexes Acted as Potential KRAS G-Quadruplex DNA Stabilizer Induced DNA Damage Mediated Apoptosis to Inhibit Breast Cancer Progress. Molecules 2022, 27, 3046. [Google Scholar] [CrossRef]
- Zeng, L.; Yuan, C.; Shu, J.; Qian, J.; Wu, Q.; Chen, Y.; Wu, R.; Ouyang, X.; Li, Y.; Mei, W. Arene Ru(II) Complexes with Difluorinated Ligands Act as Potential Inducers of S-Phase Arrest via the Stabilization of c-myc G-Quadruplex DNA. Molecules 2022, 27, 1897. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.; Sheng, Y.; Li, W.; Wang, J.; Ma, Y.; Du, B.; Tang, Y. Erianin, a novel dibenzyl compound in Dendrobium extract, inhibits bladder cancer cell growth via the mitochondrial apoptosis and JNK pathways. Toxicol. Appl. Pharm. 2019, 371, 41–54. [Google Scholar] [CrossRef]
- Gong, Y.J.; Zhang, X.B.; Mao, G.J.; Su, L.; Meng, H.M.; Tan, W.; Feng, S.; Zhang, G. A unique approach toward near-infrared fluorescent probes for bioimaging with remarkably enhanced contrast. Chem. Sci. 2016, 7, 2275–2285. [Google Scholar] [CrossRef] [PubMed]
- Huang, Y.-Y.; Tian, Y.; Liu, X.-Q.; Niu, Z.; Yang, Q.-Z.; Ramamurthy, V.; Tung, C.-H.; Chen, Y.-Z.; Wu, L.-Z. Luminescent supramolecular polymer nanoparticles for ratiometric hypoxia sensing, imaging and therapy. Mater. Chem. Front. 2018, 2, 1893–1899. [Google Scholar] [CrossRef]
- Chen, Z.F.; Liu, Y.C.; Liu, L.M.; Wang, H.S.; Qin, S.H.; Wang, B.L.; Bian, H.D.; Yang, B.; Fun, H.K.; Liu, H.G.; et al. Potential new inorganic antitumour agents from combining the anticancer traditional Chinese medicine (TCM) liriodenine with metal ions, and DNA binding studies. Dalton Trans. 2009, 262–272. [Google Scholar] [CrossRef]
- Andrezálová, L.; Plšíková, J.; Janočková, J.; Koňariková, K.; Žitňanová, I.; Kohútová, M.; Kožurková, M. DNA/BSA binding ability and genotoxic effect of mono- and binuclear copper (II) complexes containing a Schiff base derived from salicylaldehyde and D, L-glutamic acid. J. Organomet. Chem. 2017, 827, 67–77. [Google Scholar] [CrossRef]
- Wang, C.; Carter-Cooper, B.; Du, Y.; Zhou, J.; Saeed, M.A.; Liu, J.; Guo, M.; Roembke, B.; Mikek, C.; Lewis, E.A.; et al. Alkyne-substituted diminazene as G-quadruplex binders with anticancer activities. Eur. J. Med. Chem. 2016, 118, 266–275. [Google Scholar] [CrossRef]
- Gluszynska, A.; Juskowiak, B.; Kuta-Siejkowska, M.; Hoffmann, M.; Haider, S. Carbazole ligands as c-myc G-quadruplex binders. Int. J. Biol. Macromol. 2018, 114, 479–490. [Google Scholar] [CrossRef]









Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, Z.; Liu, W.; Li, G.; Wang, J.; Zhao, B.; Huang, P.; Mei, W. Ruthenium(II) Complexes Coupled by Erianin via a Flexible Carbon Chain as a Potential Stabilizer of c-myc G-Quadruplex DNA. Molecules 2023, 28, 1529. https://doi.org/10.3390/molecules28041529
Wang Z, Liu W, Li G, Wang J, Zhao B, Huang P, Mei W. Ruthenium(II) Complexes Coupled by Erianin via a Flexible Carbon Chain as a Potential Stabilizer of c-myc G-Quadruplex DNA. Molecules. 2023; 28(4):1529. https://doi.org/10.3390/molecules28041529
Chicago/Turabian StyleWang, Zhixiang, Wentao Liu, Guohu Li, Jiacheng Wang, Bin Zhao, Peishan Huang, and Wenjie Mei. 2023. "Ruthenium(II) Complexes Coupled by Erianin via a Flexible Carbon Chain as a Potential Stabilizer of c-myc G-Quadruplex DNA" Molecules 28, no. 4: 1529. https://doi.org/10.3390/molecules28041529
APA StyleWang, Z., Liu, W., Li, G., Wang, J., Zhao, B., Huang, P., & Mei, W. (2023). Ruthenium(II) Complexes Coupled by Erianin via a Flexible Carbon Chain as a Potential Stabilizer of c-myc G-Quadruplex DNA. Molecules, 28(4), 1529. https://doi.org/10.3390/molecules28041529