Microfluidic Technologies for Synthetic Biology
Abstract
:1. Introduction
1.1. Synthetic Biology
1.2. Microfluidics
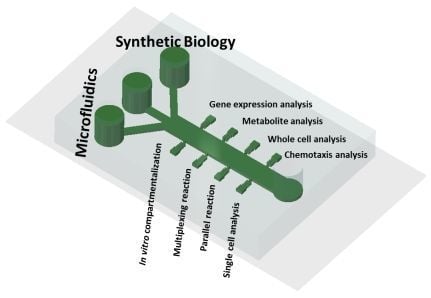
1.3. Blending Microfluidics with Synthetic Biology
2. Gene Expression and Regulation
3. Metabolite Analysis
3.1. Off-Chip Detection
3.2. On-Chip Detection
4. Whole-Cell Analysis
5. Future Perspectives
6. Conclusions
Acknowledgments
References
- Gulati, S; Rouilly, V; Niu, X; Chappell, J; Kitney, RI; Edel, JB; Freemont, PS; de Mello, AJ. Opportunities for microfluidic technologies in synthetic biology. J. R. Soc. Interface 2009, 6, S493–S506. [Google Scholar]
- Khalil, AS; Collins, JJ. Synthetic biology: Applications come of age. Nat. Rev. Genet 2010, 11, 367–379. [Google Scholar]
- Andrianantoandro, E; Basu, S; Karig, DK; Weiss, R. Synthetic biology: New engineering rules for an emerging discipline. Mol Syst Biol 2006, 2, 20060028. [Google Scholar]
- Endy, D. Foundations for engineering biology. Nature 2005, 438, 449–453. [Google Scholar]
- Breslauer, DN; Lee, PJ; Lee, LP. Microfluidics-based systems biology. Mol. Biosyst 2006, 2, 97–112. [Google Scholar]
- Bennett, MR; Hasty, J. Microfluidic devices for measuring gene network dynamics in single cells. Nat. Rev. Genet 2009, 10, 628–638. [Google Scholar]
- Elowitz, MB; Levine, AJ; Siggia, ED; Swain, PS. Stochastic gene expression in a single cell. Science 2002, 297, 1183–1186. [Google Scholar]
- Locke, JCW; Elowitz, MB. Using movies to analyse gene circuit dynamics in single cells. Nat. Rev. Micro 2009, 7, 383–392. [Google Scholar]
- Szita, N; Polizzi, K; Jaccard, N; Baganz, F. Microfluidic approaches for systems and synthetic biology. Curr. Opin. Biotechnol 2010, 21, 517–523. [Google Scholar]
- Yi, C; Li, C-W; Ji, S; Yang, M. Microfluidics technology for manipulation and analysis of biological cells. Anal. Chim. Acta 2006, 560, 1–23. [Google Scholar]
- Ozkan, M; Wang, M; Ozkan, C; Flynn, R; Esener, S. Optical manipulation of objects and biological cells in microfluidic devices. Biomed. Microdevices 2003, 5, 61–67. [Google Scholar]
- Anderson, JR; Chiu, DT; Jackman, RJ; Cherniavskaya, O; McDonald, JC; Wu, H; Whitesides, SH; Whitesides, GM. Fabrication of topologically complex three-dimensional microfluidic systems in PDMS by rapid prototyping. Anal. Chem 2000, 72, 3158–3164. [Google Scholar]
- Jeon, NL; Dertinger, SKW; Chiu, DT; Choi, IS; Stroock, AD; Whitesides, GM. Generation of solution and surface gradients using microfluidic systems. Langmuir 2000, 16, 8311–8316. [Google Scholar]
- Li, L; Du, W; Ismagilov, RF. Multiparameter screening on slipchip used for nanoliter protein crystallization combining free interface diffusion and microbatch methods. J. Am. Chem. Soc 2009, 132, 112–119. [Google Scholar]
- Shen, F; Du, W; Davydova, EK; Karymov, MA; Pandey, J; Ismagilov, RF. Nanoliter multiplex PCR arrays on a slipchip. Anal. Chem 2010, 82, 4606–4612. [Google Scholar]
- Wakamoto, Y; Umehara, S; Matsumura, K; Inoue, I; Yasuda, K. Development of non-destructive, non-contact single-cell based differential cell assay using on-chip microcultivation and optical tweezers. Sens. Actuators B 2003, 96, 693–700. [Google Scholar]
- Mao, H; Cremer, PS; Manson, MD. A sensitive, versatile microfluidic assay for bacterial chemotaxis. Proc. Natl. Acad. Sci. USA 2003, 100, 5449–5454. [Google Scholar]
- Eric, Y; Hal, A. Synthetic biology: Tools to design, build, and optimize cellular processes. J. Biomed. Biotech 130781.
- Davidsson, R; Johansson, B; Passoth, V; Bengtsson, M; Laurell, T; Emneus, J. Microfluidic biosensing systems Part II. Monitoring the dynamic production of glucose and ethanol from microchip-immobilised yeast cells using enzymatic chemiluminescent μ-biosensors. Lab Chip 2004, 4, 488–494. [Google Scholar]
- Huebner, A; Srisa-Art, M; Holt, D; Abell, C; Hollfelder, F; de Mello, AJ; Edel, JB. Quantitative detection of protein expression in single cells using droplet microfluidics. Chem Commun 2007, 1218–1220. [Google Scholar]
- Vyawahare, S; Griffiths, AD; Merten, CA. Miniaturization and parallelization of biological and chemical assays in microfluidic devices. Chem. Biol 2010, 17, 1052–1065. [Google Scholar]
- Casadevall i Solvas, X; deMello, A. Droplet microfluidics: Recent developments and future applications. Chem. Commun 2011, 47, 1936–1942. [Google Scholar]
- Shim, JU; Olguin, LF; Whyte, G; Scott, D; Babtie, A; Abell, C; Huck, WTS; Hollfelder, F. Simultaneous determination of gene expression and enzymatic activity in individual bacterial cells in microdroplet compartments. J. Am. Chem. Soc 2009, 131, 15251–15256. [Google Scholar]
- King, KR; Wang, SH; Irimia, D; Jayaraman, A; Toner, M; Yarmush, ML. A high-throughput microfluidic real-time gene expression living cell array. Lab Chip 2007, 7, 77–85. [Google Scholar]
- Thompson, DM; King, KR; Wieder, KJ; Toner, M; Yarmush, ML; Jayaraman, A. Dynamic gene expression profiling using a microfabricated living cell array. Anal. Chem 2004, 76, 4098–4103. [Google Scholar]
- Ingham, CJ; Sprenkels, A; Bomer, J; Molenaar, D; van den Berg, A; Vlieg, JETV; de Vos, WM. The micro-petri dish, a million-well growth chip for the culture and high-throughput screening of microorganisms. Proc. Natl. Acad. Sci. USA 2007, 104, 18217–18222. [Google Scholar]
- Ryley, J; Pereira-Smith, OM. Microfluidics device for single cell gene expression analysis in Saccharomyces cerevisiae. Yeast 2006, 23, 1065–1073. [Google Scholar]
- Jiang, W; Kim, BYS; Rutka, JT; Chan, WCW. Nanoparticle-mediated cellular response is size-dependent. Nat. Nanotech 2008, 3, 145–150. [Google Scholar]
- Cho, YK; Shin, H; Lee, SK; Kim, T. Current application of micro/nano-interfaces to stimulate and analyze cellular responses. Ann. Biomed. Eng 2010, 38, 2056–2067. [Google Scholar]
- Balaban, NQ; Merrin, J; Chait, R; Kowalik, L; Leibler, S. Bacterial persistence as a phenotypic switch. Science 2004, 305, 1622–1625. [Google Scholar]
- Boedicker, JQ; Vincent, ME; Ismagilov, RF. Microfluidic confinement of single cells of bacteria in small volumes initiates high-density behavior of quorum sensing and growth and reveals its variability. Angew. Chem. Int. Ed 2009, 48, 5908–5911. [Google Scholar]
- Hatch, A; Kamholz, AE; Hawkins, KR; Munson, MS; Schilling, EA; Weigl, BH; Yager, P. A rapid diffusion immunoassay in a T-sensor. Nat. Biotech 2001, 19, 461–465. [Google Scholar]
- Frasch, M; Hoey, T; Rushlow, C; Doyle, H; Levine, M. Characterization and localization of the even-skipped protein of Drosophila. EMBO J 1987, 6, 749–759. [Google Scholar]
- Ashe, HL; Briscoe, J. The interpretation of morphogen gradients. Development 2006, 133, 385–394. [Google Scholar]
- Lander, AD. Morpheus unbound: Reimagining the morphogen gradient. Cell 2007, 128, 245–256. [Google Scholar]
- Gurdon, JB; Bourillot, PY. Morphogen gradient interpretation. Nature 2001, 413, 797–803. [Google Scholar]
- Charvin, G; Cross, FR; Siggia, ED. A microfluidic device for temporally controlled gene expression and long-term fluorescent imaging in unperturbed dividing yeast cells. PLoS ONE 2008, 3, e1468. [Google Scholar]
- Park, J; Bansal, T; Pinelis, M; Maharbiz, MM. A microsystem for sensing and patterning oxidative microgradients during cell culture. Lab Chip 2006, 6, 611–622. [Google Scholar]
- Blow, N. Biochemistry’s new look. Nature 2008, 455, 697–700. [Google Scholar]
- Kraly, JR; Holcomb, RE; Guan, Q; Henry, CS. Review: Microfluidic applications in metabolomics and metabolic profiling. Anal. Chem. Acta 2009, 653, 23–35. [Google Scholar]
- Theodoridis, G; Gika, HG; Wilson, ID. LC-MS-based methodology for global metabolite profiling in metabonomics/metabolomics. Trends Anal. Chem 2008, 27, 251–260. [Google Scholar]
- Amantonico, A; Oh, JY; Sobek, J; Heinemann, M; Zenobi, R. Mass spectrometric method for analyzing metabolites in yeast with single cell sensitivity. Angew. Chem. Int. Ed 2008, 47, 5382–5385. [Google Scholar]
- Fidalgo, LM; Whyte, G; Ruotolo, BT; Benesch, JLP; Stengel, F; Abell, C; Robinson, CV; Huck, WTS. Coupling microdroplet microreactors with mass spectrometry: Reading the contents of single droplets online. Angew. Chem. Int. Ed 2009, 48, 3665–3668. [Google Scholar]
- Gao, D; Wei, HB; Guo, GS; Lin, JM. Microfluidic cell culture and metabolism detection with electrospray ionization quadrupole time-of-flight mass spectrometer. Anal. Chem 2010, 82, 5679–5685. [Google Scholar]
- Lin, YQ; Schiavo, S; Orjala, J; Vouros, P; Kautz, R. Microscale LC-MS-NMR platform applied to the identification of active cyanobacterial metabolites. Anal. Chem 2008, 80, 8045–8054. [Google Scholar]
- Lonigro, SL; Valerio, F; de Angelis, M; de Bellis, P; Lavermicocca, P. Microfluidic technology applied to cell-wall protein analysis of olive related lactic acid bacteria. Int. J. Food Microbiol 2009, 130, 6–11. [Google Scholar]
- Cheng, W; Klauke, N; Sedgwick, H; Smith, GL; Cooper, JM. Metabolic monitoring of the electrically stimulated single heart cell within a microfluidic platform. Lab Chip 2006, 6, 1424–1431. [Google Scholar]
- Liu, BF; Ozaki, M; Hisamoto, H; Luo, QM; Utsumi, Y; Hattori, T; Terabe, S. Microfluidic chip toward cellular ATP and ATP-conjugated metabolic analysis with bioluminescence detection. Anal. Chem 2005, 77, 573–578. [Google Scholar]
- Clark, AM; Sousa, KM; Jennings, C; MacDougald, OA; Kennedy, RT. Continuous-flow enzyme assay on a microfluidic chip for monitoring glycerol secretion from cultured adipocytes. Anal. Chem 2009, 81, 2350–2356. [Google Scholar]
- Urbanski, JP; Johnson, MT; Craig, DD; Potter, DL; Gardner, DK; Thorsen, T. Noninvasive metabolic profiling using microfluidics for analysis of single preimplantation embryos. Anal. Chem 2008, 80, 6500–6507. [Google Scholar]
- Huebner, A; Olguin, LF; Bratton, D; Whyte, G; Huck, WTS; de Mello, AJ; Edel, JB; Abell, C; Hollfelder, F. Development of quantitative cell-based enzyme assays in microdroplets. Anal. Chem 2008, 80, 3890–3896. [Google Scholar]
- Svobodova, J; Mathur, S; Muck, A; Letzel, T; Svatos, A. Microchip-ESI-MS determination of dissociation constant of the lysozyme-NAG(3) complex. Electrophoresis 2010, 31, 2680–2685. [Google Scholar]
- Reichmuth, DS; Shepodd, TJ; Kirby, BJ. Microchip HPLC of peptides and proteins. Anal. Chem 2005, 77, 2997–3000. [Google Scholar]
- Kim, HJ; Boedicker, JQ; Choi, JW; Ismagilov, RF. Defined spatial structure stabilizes a synthetic multispecies bacterial community. Proc. Natl. Acad. Sci. USA 2008, 105, 18188–18193. [Google Scholar]
- Nichols, D; Cahoon, N; Trakhtenberg, EM; Pham, L; Mehta, A; Belanger, A; Kanigan, T; Lewis, K; Epstein, SS. Use of Ichip for high-throughput in situ cultivation of “uncultivable” microbial species. Appl. Environ. Microbiol 2010, 76, 2445–2450. [Google Scholar]
- Ashcroft, RG; Lopez, PA. Commercial high speed machines open new opportunities in high throughput flow cytometry (HTFC). J. Immunol. Methods 2000, 243, 13–24. [Google Scholar]
- Wang, MM; Tu, E; Raymond, DE; Yang, JM; Zhang, HC; Hagen, N; Dees, B; Mercer, EM; Forster, AH; Kariv, I; Marchand, PJ; Butler, WF. Microfluidic sorting of mammalian cells by optical force switching. Nat. Biotech 2005, 23, 83–87. [Google Scholar]
- Fu, AY; Spence, C; Scherer, A; Arnold, FH; Quake, SR. A microfabricated fluorescence-activated cell sorter. Nat. Biotech 1999, 17, 1109–1111. [Google Scholar]
- Han, KH; Frazier, AB. Paramagnetic capture mode magnetophoretic microseparator for high efficiency blood cell separations. Lab Chip 2006, 6, 265–273. [Google Scholar]
- Inglis, DW; Riehn, R; Austin, RH; Sturm, JC. Continuous microfluidic immunomagnetic cell separation. Appl. Phys. Lett 2004, 85, 5093–5095. [Google Scholar]
- Pamme, N; Wilhelm, C. Continuous sorting of magnetic cells via on-chip free-flow magnetophoresis. Lab Chip 2006, 6, 974–980. [Google Scholar]
- Thiel, A; Scheffold, A; Radbruch, A. Immunomagnetic cell sorting-pushing the limits. Immunotechnology 1998, 4, 89–96. [Google Scholar]
- Davis, JA; Inglis, DW; Morton, KJ; Lawrence, DA; Huang, LR; Chou, SY; Sturm, JC; Austin, RH. Deterministic hydrodynamics: Taking blood apart. Proc. Natl. Acad. Sci. USA 2006, 103, 14779–14784. [Google Scholar]
- Sethu, P; Sin, A; Toner, M. Microfluidic diffusive filter for apheresis (leukapheresis). Lab Chip 2006, 6, 83–89. [Google Scholar]
- Choi, S; Song, S; Choi, C; Park, JK. Continuous blood cell separation by hydrophoretic filtration. Lab Chip 2007, 7, 1532–1538. [Google Scholar]
- Choi, S; Song, S; Choi, C; Park, JK. Microfluidic self-sorting of mammalian cells to achieve cell cycle synchrony by hydrophoresis. Anal. Chem 2009, 81, 1964–1968. [Google Scholar]
- Huang, LR; Cox, EC; Austin, RH; Sturm, JC. Continuous particle separation through deterministic lateral displacement. Science 2004, 304, 987–990. [Google Scholar]
- Kim, M; Kim, T. Diffusion-based and long-range concentration gradients of multiple chemicals for bacterial chemotaxis assays. Anal. Chem 2010, 82, 9401–9409. [Google Scholar]
- Groisman, A; Lobo, C; Cho, HJ; Campbell, JK; Dufour, YS; Stevens, AM; Levchenko, A. A microfluidic chemostat for experiments with bacterial and yeast cells. Nat. Methods 2005, 2, 685–689. [Google Scholar]
- Lee, WC; Bhagat, AAS; Huang, S; Vliet, KJV; Han, J; Lim, CT. High-throughput cell cycle synchronization using inertial forces in spiral microchannels. Lab Chip 2007, 11, 1359–1367. [Google Scholar]
- Gibson, DG; Glass, JI; Lartigue, C; Noskov, VN; Chuang, RY; Algire, MA; Benders, GA; Montague, MG; Ma, L; Moodie, MM; Merryman, C; Vashee, S; Krishnakumar, R; Assad-Garcia, N; Andrews-Pfannkoch, C; Denisova, EA; Young, L; Qi, ZQ; Segall-Shapiro, TH; Calvey, CH; Parmar, PP; Hutchison, CA; Smith, HO; Venter, JC. Creation of a bacterial cell controlled by a chemically synthesized genome. Science 2010, 329, 52–56. [Google Scholar]
- Wang, HH; Isaacs, FJ; Carr, PA; Sun, ZZ; Xu, G; Forest, CR; Church, GM. Programming cells by multiplex genome engineering and accelerated evolution. Nature 2009, 460, 894–898. [Google Scholar]
- Haseloff, J; Ajioka, J. Synthetic biology: History, challenges and prospects. J. R. Soc. Interface 2009, 6, S389–S391. [Google Scholar]
- Morange, M. A new revolution? The place of systems biology and synthetic biology in the history of biology. EMBO Rep 2009, 10, S50–S53. [Google Scholar]
- Khandurina, J; McKnight, TE; Jacobson, SC; Waters, LC; Foote, RS; Ramsey, JM. Integrated system for rapid PCR-based DNA analysis in microfluidic devices. Anal. Chem 2000, 72, 2995–3000. [Google Scholar]
- Spurgeon, SL; Jones, RC; Ramakrishnan, R. High throughput gene expression measurement with real time PCR in a microfluidic dynamic array. PLoS ONE 2008, 3, e1662. [Google Scholar]
- Paegel, BM; Blazej, RG; Mathies, RA. Microfluidic devices for DNA sequencing: Sample preparation and electrophoretic analysis. Curr. Opin. Biotechnol 2003, 14, 42–50. [Google Scholar]
- Bau, S; Schracke, N; Kranzle, M; Wu, H; Stahler, P; Hoheisel, J; Beier, M; Summerer, D. Targeted next-generation sequencing by specific capture of multiple genomic loci using low-volume microfluidic DNA arrays. Anal. Bioanal. Chem 2009, 393, 171–175. [Google Scholar]
- Brouzes, E; Medkova, M; Savenelli, N; Marran, D; Twardowski, M; Hutchison, JB; Rothberg, JM; Link, DR; Perrimon, N; Samuels, ML. Droplet microfluidic technology for single-cell high-throughput screening. Proc. Natl. Acad. Sci. USA 2009, 106, 14195–14200. [Google Scholar]
- Churski, K; Korczyk, P; Garstecki, P. High-throughput automated droplet microfluidic system for screening of reaction conditions. Lab Chip 2010, 10, 816–818. [Google Scholar]
- Upadhyaya, S; Selvaganapathy, PR. Microfluidic devices for cell based high throughput screening. Lab Chip 2010, 10, 341–348. [Google Scholar]
- Villa-Diaz, LG; Torisawa, YS; Uchida, T; Ding, J; Nogueira-De-Souza, NC; O’Shea, KS; Takayama, S; Smith, GD. Microfluidic culture of single human embryonic stem cell colonies. Lab Chip 2009, 9, 1749–1755. [Google Scholar]
- Takayama, S. Microfluidic engineering of stem cell niches and 3D tissue models. In Vitro Cell Dev. Biol. Anim 2010, 46, S77. [Google Scholar]
- Wang, Y; Chen, ZZ; Li, QL. Microfluidic techniques for dynamic single-cell analysis. Microchim. Acta 2010, 168, 177–195. [Google Scholar]
- Choi, WS; Ha, D; Park, S; Kim, T. Synthetic multicellular cell-to-cell communication in inkjet printed bacterial cell systems. Biomaterials 2011, 32, 2500–2507. [Google Scholar]
- Lee, SH; Heinz, AJ; Shin, S; Jung, YG; Choi, SE; Park, W; Roe, JH; Kwon, S. Capillary based patterning of cellular communities in laterally open channels. Anal. Chem 2010, 82, 2900–2906. [Google Scholar]
- Bharadwaj, R; Wong, A; Knierim, B; Singh, S; Holmes, BM; Auer, M; Simmons, BA; Adams, PD; Singh, AK. High-throughput enzymatic hydrolysis of lignocellulosic biomass via in-situ regeneration. Bioresour. Technol 2011, 102, 1329–1337. [Google Scholar]
- Bharadwaj, R; Chen, ZW; Datta, S; Holmes, BM; Sapra, R; Simmons, BA; Adams, PD; Singh, AK. Microfluidic glycosyl hydrolase screening for biomass-to-biofuel conversion. Anal. Chem 2010, 82, 9513–9520. [Google Scholar]
- Balagadde, FK; You, LC; Hansen, CL; Arnold, FH; Quake, SR. Long-term monitoring of bacteria undergoing programmed population control in a microchemostat. Science 2005, 309, 137–140. [Google Scholar]




| Microfluidic Device | Potential Application in Synthetic Biology |
|---|---|
| Device with array of cells | Parallel reaction, gene expression analysis at the single-cell level |
| Device with switchable valves | The study of dynamics of gene regulation, automation |
| Chemical concentration gradient generators | Chemotaxis analysis, quorum sensing analysis, toxicity analysis |
| Microfluidic bioreactor | Evolutionary adaptation through long-term culture, multiplexing, bacterial growth, quantification of bacterial cells |
| Droplet-based microfluidics | Spatially separated parallel reaction, multiplexing, functionbased high-throughput screening of engineered enzymes |
| In vitro compartmentalization | Parallel reaction, analysis of bacterial community structure, synthetic consortium analysis |
© 2011 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Vinuselvi, P.; Park, S.; Kim, M.; Park, J.M.; Kim, T.; Lee, S.K. Microfluidic Technologies for Synthetic Biology. Int. J. Mol. Sci. 2011, 12, 3576-3593. https://doi.org/10.3390/ijms12063576
Vinuselvi P, Park S, Kim M, Park JM, Kim T, Lee SK. Microfluidic Technologies for Synthetic Biology. International Journal of Molecular Sciences. 2011; 12(6):3576-3593. https://doi.org/10.3390/ijms12063576
Chicago/Turabian StyleVinuselvi, Parisutham, Seongyong Park, Minseok Kim, Jung Min Park, Taesung Kim, and Sung Kuk Lee. 2011. "Microfluidic Technologies for Synthetic Biology" International Journal of Molecular Sciences 12, no. 6: 3576-3593. https://doi.org/10.3390/ijms12063576
APA StyleVinuselvi, P., Park, S., Kim, M., Park, J. M., Kim, T., & Lee, S. K. (2011). Microfluidic Technologies for Synthetic Biology. International Journal of Molecular Sciences, 12(6), 3576-3593. https://doi.org/10.3390/ijms12063576