Proteomic Analysis of Albumins and Globulins from Wheat Variety Chinese Spring and Its Fine Deletion Line 3BS-8
Abstract
:1. Introduction
2. Results and Discussion
2.1. Molecular Identification of 3B Fine Deletion Lines by PCR
2.2. Morphological Characters of CS and Fine Deletion Line 3BS-8
2.3. Leaf Proteome Analysis of Chinese Spring and 3BS-8
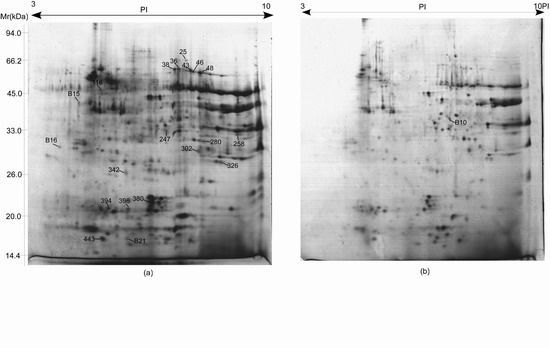
2.4. Proteome Analysis of Albumins and Globulins from Mature Seeds of CS and 3BS-8
3. Experimental Section
3.1. Plant Materials
3.2. DNA Extraction and PCR Amplification
3.3. Protein Preparation, 2-DE and Image Analysis
3.4. Protein Identification through Mass Spectrometry
4. Conclusions
Acknowledgements
References
- Badea, A.; Eudes, F.; Graf, R.J.; Laroche, A.; Gaudet, D.A.; Sadasivaiah, R.S. Phenotypic and marker-assisted evaluation of spring and winter wheat germplasm for resistance to fusarium head blight. Euphytica 2008, 164, 803–819. [Google Scholar]
- Paux, E.; Sourdille, P.; Salse, J.; Saintenac, C.; Choulet, F.; Leroy, P.; Korol, A.; Michalak, M.; Kianian, S.; Spielmeyer, W.; et al. A physical map of the 1-gigabase bread wheat chromosome 3B. Science 2008, 322, 101–104. [Google Scholar]
- Munkvold, J.D.; Greene, R.A.; Bermudez-Kandianis, C.E.; La Rota, C.M.; Edwards, H.; Sorrells, S.F.; Dake, T.; Benscher, D.; Kantety, R.; Linkiewicz, A.M.; et al. Group 3 Chromosome Bin Maps of Wheat and Their Relationship to Rice Chromosome 1. Genetics 2004, 168, 639–650. [Google Scholar]
- Sears, E.R. The aneuploids of common wheat. Res. Bull 1954, 572, 1–58. [Google Scholar]
- Ogihara, Y.; Hasegawa, K.; Tsujimoto, H. Highresolution cytological mapping of the long arm of chromosome 5A in common wheat using a series of deletion lines induced by gametocidal (Gc) genes of Aegilops speltoides. Mol. Gen. Genet 1994, 244, 253–259. [Google Scholar]
- Endo, T.R.; Gill, B.S. The deletion stocks of common wheat. J. Hered 1996, 87, 295–307. [Google Scholar]
- Endo, T.R. The gametocidal chromosome as a tool for chromosome manipulation in wheat. Chromosome Res 2007, 15, 67–75. [Google Scholar]
- Nasuda, S.; Friebe, B.; Gill, B.S. Gametocidal genes induce chromosome breakage in the interphase prior to the first mitotic cell division of the male gametophyte in wheat. Genetics 1998, 149, 1115–1124. [Google Scholar]
- Tsujimoto, H.; Noda, K. Deletion mapping by gametocidal genes in common wheat: Position of speltoid suppression (Q) and β-amylase (β-Amy-A2) genes on chromosome 5A. Genome 1990, 33, 850–853. [Google Scholar]
- Francs, C.; Thiellement, H. Chromosomal localization of structural genes and regulators in wheat by 2D electrophoresis of ditelosomic lines. Theor. Appl. Genet 1985, 71, 31–38. [Google Scholar]
- Payne, P.I.; Holt, L.M.; Jarvis, M.G.; Jackson, E.A. Two-dimensional fractionation of the endosperm proteins of bread wheat. Cereal Chem 1985, 62, 319–326. [Google Scholar]
- Islam, N.; Woo, S.H.; Tsujimoto, H.; Kawasaki, H.; Hirano, H. Proteome approaches to characterize seed storage proteins related to ditelocentric chromosomes in common wheat (Triticum aestivum L). Proteomics 2002, 2, 1146–1155. [Google Scholar]
- Islam, N.; Tsujimoto, H.; Hirano, H. Wheat proteomics: Relationship between fine chromosome deletion and protein expression. Proteomics 2003, 3, 307–316. [Google Scholar]
- Merlino, M.; Bousbata, S.; Svensson, B.; Branlard, G. Proteomic and genetic analysis of wheat endosperm albumins and globulins using deletion lines of cultivar Chinese Spring. Theor. Appl. Genet 2012. [Google Scholar] [CrossRef]
- Pierre, S.; Sukhwinder, S.; Thierry, C.; Gina, L.; Brown, G.; Georges, G.; Qi, L.; Bikram, S.G.; Philippe, D.; Alain, M.; et al. Microsatellite-based deletion bin system for the establishment of genetic-physical map relationships in wheat (Triticum aestivum L.). Funct. Integr. Genomics 2004, 4, 12–25. [Google Scholar]
- Sourdille, P.; Cadalen, T.; Gay, G.; Gill, B.; Bernard, M. Molecular and physical mapping of genes affecting awning in wheat. Plant Breed 2002, 121, 320–324. [Google Scholar]
- Watkins, A.E.; Ellerton, S. Variation and genetics of the awn in Triticum. J. Genet 1940, 40, 243–270. [Google Scholar]
- McIntosh, R.A.; Hart, G.E.; Devos, K.M.; Gale, M.D.; Rogers, W.J. Catalogue of Gene Symbols for Wheat. Available online: http://wheat.pw.usda.gov/GG2/Triticum/wgc/2008/ accessed on 12 October 2012.
- Guo, G.; Ge, P.; Ma, C.; Li, X.; Lv, D.; Wang, S.; Ma, W.; Yan, Y. Comparative proteomic analysis of salt response proteins in seedling roots of two wheat varieties. J. Proteomics 2012, 75, 1867–1885. [Google Scholar]
- Guo, G.; Lv, D.; Yan, X.; Subburaj, S.; Ge, P.; Li, X.; Hu, Y.; Yan, Y. Proteome characterization of developing grains in bread wheat cultivars (Triticum aestivum L.). BMC Plant Biol 2012. [Google Scholar] [CrossRef]
- Tatham, A.S.; Shewry, P.R. Allergens to wheat and related cereals. Clin. Exp. Allergy 2008, 38, 1712–1726. [Google Scholar]
- MacFarlane, A.J.; Burghardt, K.M.; Kelly, J.; Simell, T.; Simell, O.; Altosaar, I.; Scott, F.W. A type 1 diabetes-related protein from wheat (Triticum aestivum)—cDNA clone of a wheat storage globulin, Glb1, linked to islet damage. J. Biol. Chem 2003, 278, 54–63. [Google Scholar]
- Gomez, L.; Rosa, S.M.; Gabriel, S. A family of endosperm globulins encoded by genes located in group 1 chromosomes of wheat and related species. Mol. Gen. Genet 1988, 214, 541–546. [Google Scholar]
- Evelin, L.; Charles, W.M.; Amanda, J.M.; Fraser, W.S.; Altosaar, I. Identification of three wheat globulin genes by screening a Triticum aestivum BAC genomic library with cDNA from a diabetes-associated globulin. BMC Plant Biol 2009, 93, 1–11. [Google Scholar]
- Gao, L.Y.; Wang, A.L.; Li, X.H.; Dong, K.; Wang, K.; Appels, R.; Ma, W.J.; Yan, Y.M. Wheat quality related differential expressions of albumins and globulins revealed by two-dimensional difference gel electrophoresis (2-D DIGE). J. Proteomics 2009, 73, 279–296. [Google Scholar]
- Ge, P.; Ma, C.; Wang, S.; Gao, L.; Li, X.; Guo, G.; Ma, W.; Yan, Y. Comparative proteomic analysis of grain development in two spring wheat varieties under drought stress. Anal. Bioanal. Chem 2012, 402, 1297–1313. [Google Scholar]
- Caruso, G.; Cavaliere, C.; Guarino, C.; Gubbiotti, F.P.; Laganà, A. Identification of changes in Triticum durum L. leaf proteome in response to salt stress by two-dimensional electrophoresis and MALDI-TOF mass spectrometry. Anal. Bioanal. Chem 2008, 391, 381–390. [Google Scholar]






| Spot ID | Accession no.(gi) | Protein name | Species | p-value | Protein score a | Protein score CI% | Total ion score | Total ion score CI% | Number of matching peptides | Sequence coverage % | TpI/MW (kDa) b | EpI/MW (kDa) c |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Storage proteins | ||||||||||||
| 36 | 215398470 | globulin 3 | T. aestivum | 0.002 | 375 | 100 | 258 | 100 | 19 | 35.8 | 7.78/66.65 | 7.39/60.93 |
| 38 | 215398470 | globulin 3 | T. aestivum | 0.015 | 327 | 100 | 218 | 100 | 18 | 35.54 | 7.78/66.65 | 7.27/60.75 |
| 43 | 215398470 | globulin 3 | T. aestivum | 0.047 | 251 | 100 | 184 | 100 | 14 | 23.5 | 7.78/66.65 | 7.75/60.16 |
| 46 | 215398470 | globulin 3 | T. aestivum | 0.001 | 522 | 100 | 269 | 100 | 28 | 57.0 | 7.78/66.65 | 7.83/59.66 |
| 48 | 215398470 | globulin 3 | T. aestivum | 0.025 | 704 | 100 | 466 | 100 | 28 | 49.5 | 7.78/66.65 | 8.04/59.21 |
| 394 | 215398470 | globulin 3 | T. aestivum | 0.008 | 244 | 100 | 203 | 100 | 9 | 18.7 | 7.78/66.65 | 5.26/21.39 |
| 396 | 215398470 | globulin 3 | T. aestivum | 0.001 | 116 | 100 | 78 | 99.994 | 10 | 17.5 | 7.78/66.65 | 5.86/21.13 |
| 280 | 122232330 | Avenin-like b1 | T. aestivum | 0.037 | 246 | 100 | 209 | 100 | 5 | 22.1 | 8.08/33.79 | 8.14/35.36 |
| 302 | 110341801 | globulin 1 | T. aestivum | 0.028 | 506 | 100 | 390 | 100 | 12 | 67.7 | 8.05/25.11 | 7.99/32.85 |
| 326 | 110341795 | globulin 1 | T. aestivum | 0.075 | 631 | 100 | 433 | 100 | 18 | 80.9 | 8.57/25.10 | 8.67/30.27 |
| 380 | 215398472 | globulin 3B | T. aestivum | 0.0025 | 56 | 95.298 | 45 | 99.871 | 4 | 7.74 | 7.36/57.06 | 6.57/22.39 |
| 25 | 46981764 | HMW glutenin subunit Dty10 | Ae. tauschii | 0.048 | 423 | 100 | 318 | 100 | 12 | 46.0 | 8.2/27.38 | 7.62/65.01 |
| B15 d | 171027826 | triticin | T. aestivum | 0.048 | 170 | 100 | 109 | 100 | 12 | 17.8 | 6.43/65.29 | 4.38/47.33 |
| Carbon metabolism | ||||||||||||
| 116 | 525291 | ATP synthase beta subunit | T. aestivum | 0.003 | 623 | 100 | 467 | 100 | 19 | 52.8 | 5.56/59.33 | 5.04/52.00 |
| 247 | 357134729 | glucose and ribitol dehydrogenase | Brachypodium distachyon | 0.029 | 102 | 99.993 | 57 | 99.097 | 8 | 26.4 | 9.34/37.92 | 7.02/38.24 |
| B10 d | 253783729 | glyceraldehyde-3-phosphate dehydrogenase | T. aestivum | 0.010 | 341 | 100 | 213 | 100 | 15 | 40.9 | 6.67/36.62 | 7.44/40.96 |
| Stress/defence/Det oxification | ||||||||||||
| 258 | 22001285 | peroxidase 1 | T. aestivum | 0.028 | 408 | 100 | 228 | 100 | 19 | 45.53 | 8.14/39.26 | 9.18/37.12 |
| 443 | 326513238 | Late embryogenesis abundant protein | H. vulgare | 0.025 | 185 | 100 | 93 | 100 | 9 | 78.5 | 5.57/9.97 | 5.10/17.00 |
| B21 d | 54778511 | 0.19 dimeric alpha-amylase inhibitor | T. aestivum | 0.018 | 176 | 100 | 90 | 100 | 8 | 79.83 | 6.49/13.76 | 5.84/17.00 |
| unknown | ||||||||||||
| 342 | 326529599 | predicted protein | H. vulgare | 0.027 | 167 | 100 | 154 | 100 | 5 | 7.99 | 5.59/77.22 | 5.80/27.93 |
| B16 d | 326495978 | predicted protein | H. vulgare | 0.038 | 111 | 100 | 66 | 99.926 | 5 | 55.5 | 4.21/12.68 | 3.79/33.89 |
| Spot No. | Accession No. | Protein Name | ±da | ±ppm | Start Sequence | End Sequence | Peptide Sequence | Ion Score | Protein Score C.I.% |
|---|---|---|---|---|---|---|---|---|---|
| storage proeins | |||||||||
| 25 | gi|46981764 | HMW glutenin subunit Dty10 | 0.0778 | 85 | 78 | 86 | SVAVSQVAR | 37 | 19.535 |
| 0.0647 | 58 | 45 | 54 | QVVDQQLAGR | 71 | 99.966 | |||
| 0.0563 | 43 | 34 | 44 | ELQESSLEACR | 72 | 99.975 | |||
| 0.0288 | 20 | 227 | 239 | AQQPATQLPTVCR | 75 | 99.986 | |||
| −0.1294 | −57 | 136 | 157 | QGSYYPGQASPQQPGQGQQPGK | 40 | 62.537 | |||
| 36 | gi|215398470 | globulin 3 | −0.0166 | −17 | 132 | 139 | RPYVFGPR | 44 | 83.599 |
| −0.0045 | −4 | 154 | 163 | ALRPFDEVSR | 41 | 64.854 | |||
| 0.0014 | 1 | 364 | 374 | SFHALAQHDVR | 72 | 99.974 | |||
| 0.002 | 1 | 339 | 349 | DTFNLLEQRPK | 43 | 79.682 | |||
| 38 | gi|215398470 | globulin 3 | −0.0068 | −6 | 154 | 163 | ALRPFDEVSR | 37 | 21.378 |
| 0.0028 | 2 | 364 | 374 | SFHALAQHDVR | 77 | 99.991 | |||
| 0.01 | 7 | 339 | 349 | DTFNLLEQRPK | 50 | 95.444 | |||
| 43 | gi|215398470 | globulin 3 | −0.0069 | −6 | 154 | 163 | ALRPFDEVSR | 40 | 62.03 |
| −0.0053 | −4 | 364 | 374 | SFHALAQHDVR | 61 | 99.703 | |||
| 0.0043 | 3 | 339 | 349 | DTFNLLEQRPK | 64 | 99.857 | |||
| 46 | gi|215398470 | globulin 3 | −0.019 | −16 | 154 | 163 | ALRPFDEVSR | 41 | 68.692 |
| −0.0145 | −11 | 364 | 374 | SFHALAQHDVR | 77 | 99.993 | |||
| −0.0069 | −5 | 339 | 349 | DTFNLLEQRPK | 61 | 99.725 | |||
| 0.0088 | 5 | 470 | 488 | GSAFVVPPGHPVVEIASSR | 45 | 89.119 | |||
| −0.0022 | −1 | 446 | 464 | GSGSESEEEQDQQRYETVR | 44 | 86.858 | |||
| 48 | gi|215398470 | globulin 3 | −0.0186 | −16 | 154 | 163 | ALRPFDEVSR | 37 | 29.797 |
| −0.007 | −5 | 364 | 374 | SFHALAQHDVR | 76 | 99.989 | |||
| 0.0025 | 2 | 339 | 349 | DTFNLLEQRPK | 80 | 99.996 | |||
| 0.0347 | 18 | 470 | 488 | GSAFVVPPGHPVVEIASSR | 101 | 100 | |||
| 0.032 | 14 | 446 | 464 | GSGSESEEEQDQQRYETVR | 74 | 99.983 | |||
| 0.06 | 25 | 542 | 562 | AKDQQDEGFVAGPEQQQEHER | 80 | 99.996 | |||
| 394 | gi|215398470 | globulin 3 | 0.1372 | 72 | 470 | 488 | GSAFVVPPGHPVVEIASSR | 77 | 99.994 |
| 0.1484 | 67 | 446 | 464 | GSGSESEEEQDQQRYETVR | 60 | 99.733 | |||
| 0.2031 | 84 | 542 | 562 | AKDQQDEGFVAGPEQQQEHER | 66 | 99.935 | |||
| 396 | gi|215398470 | globulin 3 | 0.0885 | 46 | 470 | 488 | GSAFVVPPGHPVVEIASSR | 61 | 99.742 |
| 280 | gi|122232330 | Avenin-like b1 | 0.069 | 55 | 203 | 212 | QLSQIPEQFR | 77 | 99.993 |
| 0.0913 | 66 | 213 | 224 | CQAIHNVAEAIR | 92 | 100 | |||
| 0.2202 | 74 | 225 | 248 | QQQPQQQWQGMYQPQQPAQHESIR | 39 | 60.794 | |||
| 302 | gi|110341801 | globulin 1 | 0.1034 | 87 | 47 | 56 | QILEQQLTGR | 74 | 99.988 |
| 0.0934 | 67 | 35 | 46 | GEVQEKPLLACR | 69 | 99.958 | |||
| 0.0948 | 63 | 99 | 111 | DYEQSMPPLGEGR | 42 | 81.431 | |||
| 0.1016 | 56 | 57 | 74 | AGEGAVGVPLFHAQWGAR | 54 | 98.836 | |||
| 380 | gi|215398472 | globulin 3B | 0.031 | 23 | 61 | 72 | HGEGGREEEQGR | 45 | 99.871 |
| B15 | gi|171027826 | triticin | 0.0056 | 3 | 61 | 77 | SQAGLTEYFDEENEQFR | 84 | 99.999 |
| Carbon metabolism | |||||||||
| 116 | gi|525291 | ATP synthase beta subunit | 0.1179 | 85 | 249 | 262 | AHGGFSVFAGVGER | 108 | 100 |
| 0.1168 | 83 | 148 | 161 | VLNTGSPITVPVGR | 72 | 99.983 | |||
| 0.114 | 76 | 336 | 349 | FTQANSEVSALLGR | 79 | 99.996 | |||
| 0.0671 | 33 | 413 | 431 | QISELGIYPAVDPLDSTSR | 53 | 98.601 | |||
| −0.0181 | −8 | 192 | 211 | EAPAFVEQATEQQILVTG | 85 | 100 | |||
| −0.0049 | −2 | 350 | 370 | IPSAVGYQPTLATDLGGLGER | 69 | 99.959 | |||
| 247 | gi|357134729 | glucose and ribitol dehydrogenase | −0.0207 | −25 | 261 | 268 | GAIVAFTR | 31 | 0 |
| 0.0351 | 29 | 97 | 109 | VALVTGGDSGIGR | 26 | 0 | |||
| B10 | gi|253783729 | glyceraldehyde-3-phosphate dehydrogenase | 0.0334 | 28 | 301 | 311 | AGIALNDHFVK | 46 | 89.006 |
| 0.149 | 68 | 274 | 293 | GIMGYVEEDLVSTDFVGDSR | 100 | 100 | |||
| 0.1816 | 82 | 274 | 293 | GIMGYVEEDLVSTDFVGDSR | 136 | 100 | |||
| Stress/defense/detoxification | |||||||||
| 258 | gi|22001285 | peroxidase 1 | 0.0365 | 37 | 62 | 71 | DIGLAAGLLR | 44 | 86.056 |
| 0.0381 | 33 | 34 | 42 | GLSFDFYRR | 43 | 82.606 | |||
| 0.0231 | 16 | 127 | 141 | GAVVSCADILALAAR | 73 | 99.982 | |||
| 443 | gi|326513238 | embryogenesis abundant protein | 0.0187 | 14 | 30 | 41 | SLEAQQNLAEGR | 72 | 99.977 |
| B21 | gi|54778511 | 0.19 dimeric alpha-amylase inhibitor | 0.0131 | 7 | 40 | 53 | ECCQQLADISEWCR | 54 | 98.884 |
| unknown | |||||||||
| 342 | gi|326529599 | predicted protein | 0.0265 | 19 | 33 | 46 | AGAAVGGQVVEKER | 90 | 100 |
| B16 | gi|326495978 | predicted protein | −0.058 | −35 | 82 | 96 | DIELVMTQAGVPRPK | 35 | 9.731 |
| Primers | Sequences |
|---|---|
| gwm493 | 5wm493esATAACTAAAACCGCG-3 5TAACTAAAACCGCG-3 ers were s |
| gwm566 | 5wm566GTCTACCCATGGGATTTG-3s 5TCTACCCATGGGATTTG-3s were |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Ma, C.-Y.; Gao, L.-Y.; Li, N.; Li, X.-H.; Ma, W.-J.; Appels, R.; Yan, Y.-M. Proteomic Analysis of Albumins and Globulins from Wheat Variety Chinese Spring and Its Fine Deletion Line 3BS-8. Int. J. Mol. Sci. 2012, 13, 13398-13413. https://doi.org/10.3390/ijms131013398
Ma C-Y, Gao L-Y, Li N, Li X-H, Ma W-J, Appels R, Yan Y-M. Proteomic Analysis of Albumins and Globulins from Wheat Variety Chinese Spring and Its Fine Deletion Line 3BS-8. International Journal of Molecular Sciences. 2012; 13(10):13398-13413. https://doi.org/10.3390/ijms131013398
Chicago/Turabian StyleMa, Chao-Ying, Li-Yan Gao, Ning Li, Xiao-Hui Li, Wu-Jun Ma, Rudi Appels, and Yue-Ming Yan. 2012. "Proteomic Analysis of Albumins and Globulins from Wheat Variety Chinese Spring and Its Fine Deletion Line 3BS-8" International Journal of Molecular Sciences 13, no. 10: 13398-13413. https://doi.org/10.3390/ijms131013398
APA StyleMa, C. -Y., Gao, L. -Y., Li, N., Li, X. -H., Ma, W. -J., Appels, R., & Yan, Y. -M. (2012). Proteomic Analysis of Albumins and Globulins from Wheat Variety Chinese Spring and Its Fine Deletion Line 3BS-8. International Journal of Molecular Sciences, 13(10), 13398-13413. https://doi.org/10.3390/ijms131013398