Analysis of the Endoplasmic Reticulum Subproteome in the Livers of Type 2 Diabetic Mice
Abstract
:1. Introduction
2. Results
2.1. Preparation and Confirmation of ER Protein Fractions
2.2. Identification and Quantification of the Hepatic ER Subproteome
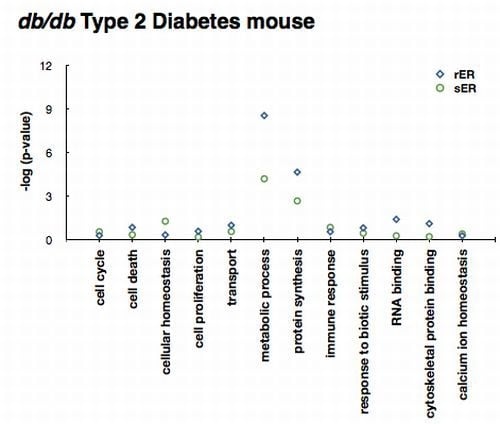
2.3. ER Dysfunction in the Type 2 Diabetic Mouse Liver
2.4. Sensing and Responding to ER Stress
3. Discussion
4. Experimental Section
4.1. Animals and Tissue Preparation
4.2. Separation and Fractionation of Organelles of Mouse Liver
4.3. SDS-PAGE and In-Gel Digestion
4.4. Nano-UPLC-MSE Tandem Mass Spectrometry and Database Search
4.5. Bioinformatic Analysis
4.6. Western Blot
5. Conclusions
Acknowledgments
- Conflict of InterestAll authors declare no conflict of interest.
References
- Vertel, B.M.; Walters, L.M.; Mills, D. Subcompartments of the endoplasmic reticulum. Semin. Cell Biol 1992, 3, 325–341. [Google Scholar]
- English, A.R.; Zurek, N.; Voeltz, G.K. Peripheral ER structure and function. Curr. Opin. Cell Biol 2009, 21, 596–602. [Google Scholar]
- Voeltz, G.K.; Rolls, M.M.; Rapoport, T.A. Structural organization of the endoplasmic reticulum. EMBO Rep 2002, 3, 944–950. [Google Scholar]
- Ozcan, L.; Tabas, I. Role of endoplasmic reticulum stress in metabolic disease and other disorders. Annu. Rev. Med 2012, 63, 317–328. [Google Scholar]
- Yoshida, H. ER stress and diseases. FEBS J 2007, 274, 630–658. [Google Scholar]
- Dara, L.; Ji, C.; Kaplowitz, N. The contribution of endoplasmic reticulum stress to liver diseases. Hepatology 2011, 53, 1752–1763. [Google Scholar]
- The Expert Committee on the Diagnosis and Classification of Diabetes Mellitus. Report of the expert committee on the diagnosis and classification of diabetes mellitus. Diabetes Care 2003, 26, S5–S20.
- Lebovitz, H.E. Type 2 diabetes: An overview. Clin. Chem 1999, 45, 1339–1345. [Google Scholar]
- Kahn, S.E.; Hull, R.L.; Utzschneider, K.M. Mechanisms linking obesity to insulin resistance and type 2 diabetes. Nature 2006, 444, 840–846. [Google Scholar]
- DeFronzo, R.A.; Simonson, D.; Ferrannini, E. Hepatic and peripheral insulin resistance: A common feature of type 2 (non-insulin-dependent) and type 1 (insulin-dependent) diabetes mellitus. Diabetologia 1982, 23, 313–319. [Google Scholar]
- Paquot, N.; Scheen, A.J.; Dirlewanger, M.; Lefebvre, P.J.; Tappy, L. Hepatic insulin resistance in obese non-diabetic subjects and in type 2 diabetic patients. Obes. Res 2002, 10, 129–134. [Google Scholar]
- Samuel, V.T.; Shulman, G.I. Mechanisms for insulin resistance: Common threads and missing links. Cell 2012, 148, 852–871. [Google Scholar]
- Flamment, M.; Hajduch, E.; Ferre, P.; Foufelle, F. New insights into ER stress-induced insulin resistance. Trends Endocrinol. MeTable 2012, 23, 381–390. [Google Scholar]
- Sundsten, T.; Ortsater, H. Proteomics in diabetes research. Mol. Cell. Endocrinol 2009, 297, 93–103. [Google Scholar]
- Sanchez, J.C.; Converset, V.; Nolan, A.; Schmid, G.; Wang, S.; Heller, M.; Sennitt, M.V.; Hochstrasser, D.F.; Cawthorne, M.A. Effect of rosiglitazone on the differential expression of obesity and insulin resistance associated proteins in lep/lep mice. Proteomics 2003, 3, 1500–1520. [Google Scholar]
- Edvardsson, U.; von Lowenhielm, H.B.; Panfilov, O.; Nystrom, A.C.; Nilsson, F.; Dahllof, B. Hepatic protein expression of lean mice and obese diabetic mice treated with peroxisome proliferator-activated receptor activators. Proteomics 2003, 3, 468–478. [Google Scholar]
- Schmid, G.M.; Converset, V.; Walter, N.; Sennitt, M.V.; Leung, K.Y.; Byers, H.; Ward, M.; Hochstrasser, D.F.; Cawthorne, M.A.; Sanchez, J.C. Effect of high-fat diet on the expression of proteins in muscle, adipose tissues, and liver of C57BL/6 mice. Proteomics 2004, 4, 2270–2282. [Google Scholar]
- Baiges, I.; Palmfeldt, J.; Blade, C.; Gregersen, N.; Arola, L. Lipogenesis is decreased by grape seed proanthocyanidins according to liver proteomics of rats fed a high fat diet. Mol. Cell. Proteomics 2010, 9, 1499–1513. [Google Scholar]
- Valle, A.; Catalan, V.; Rodriguez, A.; Rotellar, F.; Valenti, V.; Silva, C.; Salvador, J.; Fruhbeck, G.; Gomez-Ambrosi, J.; Roca, P.; Oliver, J. Identification of liver proteins altered by type 2 diabetes mellitus in obese subjects. Liver Int 2012, 32, 951–961. [Google Scholar]
- Song, Y.; Hao, Y.; Sun, A.; Li, T.; Li, W.; Guo, L.; Yan, Y.; Geng, C.; Chen, N.; Zhong, F.; et al. Sample preparation project for the subcellular proteome of mouse liver. Proteomics 2006, 6, 5269–5277. [Google Scholar]
- Song, Y.; Jiang, Y.; Ying, W.; Gong, Y.; Yan, Y.; Yang, D.; Ma, J.; Xue, X.; Zhong, F.; Wu, S.; et al. Quantitative proteomic survey of endoplasmic reticulum in mouse liver. J. Proteome Res 2010, 9, 1195–1202. [Google Scholar]
- Hummel, K.P.; Dickie, M.M.; Coleman, D.L. Diabetes, a new mutation in the mouse. Science 1966, 153, 1127–1128. [Google Scholar]
- Matsuda, T.; Kido, Y.; Uchida, T.; Kasuga, M. Reduced insulin signaling and endoplasmic reticulum stress act synergistically to deteriorate pancreatic beta cell function. Kobe J. Med. Sci 2008, 54, E114–E121. [Google Scholar]
- Gao, B.B.; Stuart, L.; Feener, E.P. Label-free quantitative analysis of one-dimensional PAGE LC/MS/MS proteome: Application on angiotensin II-stimulated smooth muscle cells secretome. Mol. Cell. Proteomics 2008, 7, 2399–2409. [Google Scholar]
- Hummasti, S.; Hotamisligil, G.S. Endoplasmic reticulum stress and inflammation in obesity and diabetes. Circ. Res 2010, 107, 579–591. [Google Scholar]
- Van der Kallen, C.J.; van Greevenbroek, M.M.; Stehouwer, C.D.; Schalkwijk, C.G. Endoplasmic reticulum stress-induced apoptosis in the development of diabetes: Is there a role for adipose tissue and liver? Apoptosis 2009, 14, 1424–1434. [Google Scholar]
- Lin, J.H.; Li, H.; Yasumura, D.; Cohen, H.R.; Zhang, C.; Panning, B.; Shokat, K.M.; Lavail, M.M.; Walter, P. IRE1 signaling affects cell fate during the unfolded protein response. Science 2007, 318, 944–949. [Google Scholar]
- Wang, Y.; Vera, L.; Fischer, W.H.; Montminy, M. The CREB coactivator CRTC2 links hepatic ER stress and fasting gluconeogenesis. Nature 2009, 460, 534–537. [Google Scholar]
- Han, M.S.; Chung, K.W.; Cheon, H.G.; Rhee, S.D.; Yoon, C.H.; Lee, M.K.; Kim, K.W.; Lee, M.S. Imatinib mesylate reduces endoplasmic reticulum stress and induces remission of diabetes in db/db mice. Diabetes 2009, 58, 329–336. [Google Scholar]
- Morand, J.P.; Macri, J.; Adeli, K. Proteomic profiling of hepatic endoplasmic reticulum-associated proteins in an animal model of insulin resistance and metabolic dyslipidemia. J. Biol. Chem 2005, 280, 17626–17633. [Google Scholar]
- Ni, M.; Lee, A.S. ER chaperones in mammalian development and human diseases. FEBS Lett 2007, 581, 3641–3651. [Google Scholar]
- Malhi, H.; Kaufman, R.J. Endoplasmic reticulum stress in liver disease. J. Hepatol 2011, 54, 795–809. [Google Scholar]
- Yamagishi, N.; Ueda, T.; Mori, A.; Saito, Y.; Hatayama, T. Decreased expression of endoplasmic reticulum chaperone GRP78 in liver of diabetic mice. Biochem. Biophys. Res. Commun 2012, 417, 364–370. [Google Scholar]
- Li, T.; Woods, J.S. Cloning, expression, and biochemical properties of CPOX4, a genetic variant of coproporphyrinogen oxidase that affects susceptibility to mercury toxicity in humans. Toxicol. Sci 2009, 109, 228–236. [Google Scholar]
- Wang, W.A.; Groenendyk, J.; Michalak, M. Calreticulin signaling in health and disease. Int. J. Biochem. Cell Biol 2012, 44, 842–846. [Google Scholar]
- Williams, D.B. Beyond lectins: The calnexin/calreticulin chaperone system of the endoplasmic reticulum. J. Cell Sci 2006, 119, 615–623. [Google Scholar]
- Christensen, A.; Svensson, K.; Persson, S.; Jung, J.; Michalak, M.; Widell, S.; Sommarin, M. Functional characterization of Arabidopsis calreticulin1a: A key alleviator of endoplasmic reticulum stress. Plant Cell Physiol 2008, 49, 912–924. [Google Scholar]
- Huang, J.; Viswakarma, N.; Yu, S.; Jia, Y.; Bai, L.; Vluggens, A.; Cherkaoui-Malki, M.; Khan, M.; Singh, I.; Yang, G.; et al. Progressive endoplasmic reticulum stress contributes to hepatocarcinogenesis in fatty acyl-CoA oxidase 1-deficient mice. Am. J. Pathol 2011, 179, 703–713. [Google Scholar]
- Xie, W.D.; Wang, H.; Zhang, J.F.; Li, J.N.; Can, Y.; Qing, L.; Kung, H.F.; Zhang, Y.O. Enhanced peroxisomal β-oxidation metabolism in visceral adipose tissues of high-fat diet-fed obesity-resistant C57BL/6 mice. Exp. Ther. Med 2011, 2, 309–315. [Google Scholar]
- Wang, B.; Zhou, S.F. Synthetic and natural compounds that interact with human cytochrome P450 1A2 and implications in drug development. Curr. Med. Chem 2009, 16, 4066–4218. [Google Scholar]
- Shimojo, N. Cytochrome P450 changes in rats with streptozocin-induced diabetes. Int. J. Biochem 1994, 26, 1261–1268. [Google Scholar]
- Wang, Z.; Hall, S.D.; Maya, J.F.; Li, L.; Asghar, A.; Gorski, J.C. Diabetes mellitus increases the in vivo activity of cytochrome P450 2E1 in humans. Br. J. Clin. Pharmacol 2003, 55, 77–85. [Google Scholar]
- Ramaiah, S.K.; Apte, U.; Mehendale, H.M. Cytochrome P4502E1 induction increases thioacetamide liver injury in diet-restricted rats. Drug Metab. Dispos 2001, 29, 1088–1095. [Google Scholar]
- Moon, Y.J.; Kwon, J.; Yun, S.H.; Lim, H.L.; Kim, M.S.; Kang, S.G.; Lee, J.H.; Choi, J.S.; Kim, S.I.; Chung, Y.H. Proteome Analyses of Hydrogen-producing Hyperthermophilic Archaeon Thermococcus onnurineus NA1 in Different One-carbon Substrate Culture Conditions. Mol. Cell. Proteomics 2012, 11. [Google Scholar] [CrossRef]
- Ingenuity Pathway Analysis. Available online: http://www.ingenuity.com accessed on 27 September 2012.
- GOfact. Available online: http://61.50.138.118/gofact/cgi/gofact2009.cgi accessed on 27 September 2012.
- Park, E.C.; Cho, G.S.; Kim, G.H.; Choi, S.C.; Han, J.K. The involvement of Eph-Ephrin signaling in tissue separation and convergence during Xenopus gastrulation movements. Dev. Biol 2011, 350, 441–450. [Google Scholar]





| Ratio | C57 (rER/sER) | db/db (rER/sER) | rER (db/db/C57) | sER (db/db/C57) | ||||
|---|---|---|---|---|---|---|---|---|
| T_Pro * | U_Pro ** | T_Pro | U_Pro | T_Pro | U_Pro | T_Pro | U_Pro | |
| Ratio ≥ 5 | 360 | 174 | 251 | 118 | 269 | 133 | 276 | 143 |
| 5 > Ratio ≥ 1.5 | 102 | 40 | 69 | 30 | 122 | 40 | 108 | 42 |
| 1.5 > Ratio > 0.67 | 319 | 113 | 362 | 146 | 282 | 113 | 297 | 91 |
| 0.2 < Ratio ≤ 0.67 | 63 | 23 | 33 | 15 | 76 | 26 | 66 | 27 |
| Ratio ≤ 0.2 | 301 | 143 | 221 | 106 | 325 | 165 | 313 | 159 |
| Subtotal | 1145 | 493 | 936 | 415 | 1074 | 477 | 1060 | 462 |
| Total (T_Pro/U_Pro) | 1584/685 | |||||||
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Park, E.C.; Kim, G.-H.; Yun, S.-H.; Lim, H.L.; Hong, Y.; Kwon, S.-O.; Kwon, J.; Chung, Y.-H.; Kim, S.I. Analysis of the Endoplasmic Reticulum Subproteome in the Livers of Type 2 Diabetic Mice. Int. J. Mol. Sci. 2012, 13, 17230-17243. https://doi.org/10.3390/ijms131217230
Park EC, Kim G-H, Yun S-H, Lim HL, Hong Y, Kwon S-O, Kwon J, Chung Y-H, Kim SI. Analysis of the Endoplasmic Reticulum Subproteome in the Livers of Type 2 Diabetic Mice. International Journal of Molecular Sciences. 2012; 13(12):17230-17243. https://doi.org/10.3390/ijms131217230
Chicago/Turabian StylePark, Edmond Changkyun, Gun-Hwa Kim, Sung-Ho Yun, Hye Li Lim, Yeonhee Hong, Sang-Oh Kwon, Joseph Kwon, Young-Ho Chung, and Seung Il Kim. 2012. "Analysis of the Endoplasmic Reticulum Subproteome in the Livers of Type 2 Diabetic Mice" International Journal of Molecular Sciences 13, no. 12: 17230-17243. https://doi.org/10.3390/ijms131217230
APA StylePark, E. C., Kim, G. -H., Yun, S. -H., Lim, H. L., Hong, Y., Kwon, S. -O., Kwon, J., Chung, Y. -H., & Kim, S. I. (2012). Analysis of the Endoplasmic Reticulum Subproteome in the Livers of Type 2 Diabetic Mice. International Journal of Molecular Sciences, 13(12), 17230-17243. https://doi.org/10.3390/ijms131217230