Recent Molecular Advances on Downstream Plant Responses to Abiotic Stress
Abstract
:1. Introduction
2. Heat-Shock Proteins
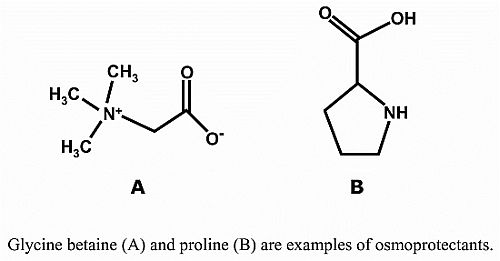
3. Osmoprotectants
3.1. Proline
3.2. Glycine Betaine
3.3. Trehalose
3.4. Other Sugar Alcohols
4. LEA Protein
5. Conclusions and Perspectives
Acknowledgements
References
- Alcázar, R.; Marco, F.; Cuevas, J.C.; Patron, M.; Ferrando, A.; Carrasco, P.; Tiburcio, A.F.; Altabella, T. Involvement of polyamines in plant response to abiotic stress. Biotechnol. Lett 2006, 28, 1867–1876. [Google Scholar]
- Flowers, T.J.; Yeo, A.R. Breeding for salinity resistance in crop plants: Where next? Aust. J. Plant Physiol 1995, 22, 875–884. [Google Scholar]
- Ahuja, I.; de Vos, R.C.H.; Bones, A.M.; Hall, R.D. Plant molecular stress responses face climate change. Trends Plant Sci 2010, 15, 664–674. [Google Scholar]
- Bhatnagar-Mathur, P.; Vadez, V.; Sharma, K.K. Transgenic approaches for abiotic stress tolerance in plants: Retrospect and prospects. Plant Cell Rep 2008, 27, 411–424. [Google Scholar]
- Mittler, R. Abiotic stress, the field environment and stress combination. Trends Plant Sci 2006, 11, 15–19. [Google Scholar]
- Fujita, M.; Fujita, Y.; Noutoshi, Y.; Takahashi, F.; Narusaka, Y.; Yamaguchi-Shinozaki, K.; Shinozaki, K. Crosstalk between abiotic and biotic stress responses: A current view from the points of convergence in the stress signaling networks. Curr. Opin. Plant Biol 2006, 9, 436–442. [Google Scholar]
- Chinnusamy, V.; Schumaker, K.; Zhu, J.-K. Molecular genetic perspectives on cross-talk and specificity in abiotic stress signalling in plants. J. Exp. Bot 2004, 55, 225–236. [Google Scholar]
- Shinozaki, K.; Yamaguchi-Shinozaki, K. Gene networks involved in drought stress response and tolerance. J. Exp. Bot 2007, 58, 221–227. [Google Scholar]
- Krasensky, J.; Jonak, C. Drought, salt, and temperature stress-induced metabolic rearrangements and regulatory networks. J. Exp. Bot 2011. [Google Scholar] [CrossRef]
- Bohnert, H.J.; Ayoubi, P.; Borchert, C.; Bressan, R.A.; Burnap, R.L.; Cushman, J.C.; Cushman, M.A.; Deyholos, M.; Fisher, R.; Galbraith, D.W.; et al. A genomic approach towards salt stress tolerance. Plant Physiol. Biochem 2001, 39, 295–311. [Google Scholar]
- Seki, M.; Narusaka, M.; Abe, H.; Kasuga, M.; Yamaguchi-Shinozaki, K.; Carninci, P.; Hayashizaki, Y.; Shinozaki, K. Monitoring the expression pattern of 1300 Arabidopsis genes under drought and cold stresses by using a full-length cDNA microarray. Plant Cell 2001, 13, 61–72. [Google Scholar]
- Zhu, T.; Budworth, P.; Han, B.; Brown, D.; Chang, H.S.; Zou, G.; Wang, X. Toward elucidating the global expression patterns of developing Arabidopsis: Parallel analysis of 8300 genes by a high-density oligonucleotide probe array. Plant. Physiol. Biochem 2001, 39, 221–242. [Google Scholar]
- Agarwal, P.K.; Agarwal, P.; Reddy, M.K.; Sopory, S.K. Role of DREB transcription factors in abiotic and biotic stress tolerance in plants. Plant Cell Rep 2001, 25, 1263–1274. [Google Scholar]
- Kreps, J.A.; Wu, Y.; Chang, H.-S.; Zhu, T.; Wang, X.; Harper, J.F. Transcriptome changes for Arabidopsis in response to salt, osmotic, and cold stress. Plant Physiol 2002, 130, 2129–2141. [Google Scholar]
- Rizhsky, L.; Liang, H.; Shuman, J.; Shulaev, V.; Davletova, S.; Mittler, R. When defense pathways collide. The response of Arabidopsis to a combination of drought and heat stress. Plant Physiol 2004, 134, 1683–1696. [Google Scholar]
- Vinocur, B.; Altman, A. Recent advances in engineering plant tolerance to abiotic stress: Achievements and limitations. Curr. Opin. Biotechnol 2005, 16, 123–132. [Google Scholar]
- Kotak, S.; Larkindale, J.; Lee, U.; Koskull-Döring, P.; Vierling, E.; Scharf, K.-D. Complexity of the heat stress response in plants. Curr. Opin. Plant Biol 2007, 10, 310–316. [Google Scholar]
- Wang, W.; Vinocur, B.; Shoseyov, O.; Altman, A. Role of plant heat shock proteins and molecular chaperones in the abiotic stress response. Trends Plant Sci 2004, 9, 244–252. [Google Scholar]
- Iba, K. Acclimative response to temperature stress in higher plants: Approaches of gene engineering for temperature tolerance. Annu. Rev. Plant Biol 2002, 53, 225–245. [Google Scholar]
- Miller, G.; Mittler, R. Could heat shock transcription factors function as hydrogen peroxide sensors in plants? Ann. Bot 2006, 98, 279–288. [Google Scholar]
- Sabehat, A.; Weiss, D.; Lurie, S. Heat-shock proteins and cross-tolerance in plants. Physiol. Plant 1998, 103, 437–441. [Google Scholar]
- Vierling, E. The roles of heat shock proteins in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol 1991, 42, 579–620. [Google Scholar]
- Baniwal, S.K.; Bharti, K.; Chan, K.Y.; Fauth, M.; Ganguli, A.; Kotak, S.; Mishra, S.K.; Nover, L.; Port, M.; Scharf, K.-D.; et al. Heat stress response in plants: A complex game with chaperones and more than twenty heat stress transcription factors. J. Biosci 2004, 29, 471–487. [Google Scholar]
- Hahn, J.S.; Hu, Z.; Thiele, D.J.; Iyer, V.R. Genome-wide analysis of the biology of stress responses through heat shock transcription factor. Mol. Cell. Biol 2004, 24, 5249–5256. [Google Scholar]
- Scharf, K.-D.; Berberich, T.; Ebersberger, I.; Nover, L. The plant heat stress transcription factor (Hsf) family: Structure, function and evolution. Biochim. Biophys. Acta 2012, 1819, 104–119. [Google Scholar]
- Giorno, F.; Wolters-Arts, M.; Grillo, S.; Scharf, K.-D.; Vriezen, W.-H.; Mariani, C. Developmental and heat stress-regulated expression of HsfA2 and small heat shock proteins in tomato anthers. J. Exp. Bot 2010, 61, 453–462. [Google Scholar]
- Hu, W.; Hu, G.; Han, B. Genome-wide survey and expression profiling of heat shock proteins and heat shock factors revealed overlapped and stress specific response under abiotic stresses in rice. Plant. Sci 2009, 176, 583–590. [Google Scholar]
- Swindell, W.R.; Huebner, M.; Weber, A.P. Transcriptional profiling of Arabidopsis heat shock proteins and transcription factors reveals extensive overlap between heat and non-heat stress response pathways. BMC Genomics 2007, 8, 1–15. [Google Scholar]
- Nishizawa-Yokoi, A.; Nosaka, R.; Hayashi, H.; Tainaka, H.; Maruta, T.; Tamoi, M.; Ikeda, M.; Ohme-Takagi, M.; Yoshimura, K.; Yabuta, Y.; et al. HsfA1d and HsfA1e involved in the transcriptional regulation of HsfA2 function as key regulators for the Hsf signaling network in response to environmental stress. Plant Cell Physiol 2011, 52, 933–945. [Google Scholar]
- Hahn, A.; Bublak, D.; Schleiff, E.; Scharf, K.-D. Crosstalk between Hsp90 and Hsp70 chaperones and heat stress transcription factors in tomato. Plant Cell 2011, 23, 741–755. [Google Scholar]
- Banti, V.; Mafessoni, F.; Loreti, E.; Alpi, A.; Perata, P. The heat-inducible transcription factor HsfA2 enhances anoxia tolerance in Arabidopsis. Plant Physiol 2010, 152, 1471–1483. [Google Scholar]
- Chauhan, H.; Khurana, N.; Agarwal, P.; Khurana, P. Heat shock factors in rice (Oryza sativa L.): Genome-wide expression analysis during reproductive development and abiotic stress. Mol. Genet. Genomics 2011, 286, 171–187. [Google Scholar]
- Frank, G.; Pressman, E.; Ophir, R.; Althan, L.; Shaked, R.; Freedman, M.; Shen, S.; Firon, N. Transcriptional profiling of maturing tomato (Solanum lycopersicum L.) microspores reveals the involvement of heat shock proteins, ROS scavengers, hormones, and sugars in the heat stress response. J. Exp. Bot 2009, 60, 3891–3908. [Google Scholar]
- Ye, S.F.; Yu, S.W.; Shu, L.B.; Wu, J.H.; Wu, A.Z.; Luo, L.J. Expression profile analysis of 9 heat shock protein genes throughout the life cycle and under abiotic stress in rice. Chin. Sci. Bull 2012, 57, 336–343. [Google Scholar]
- Park, H.-S.; Jeong, W.-J.; Kim, E.-C.; Jung, Y.; Lim, J.M.; Hwang, M.S.; Park, E.-J.; Ha, D.-S.; Choi, D.-W. Heat shock protein gene family of the Porphyra seriata and enhancement of heat stress tolerance by PsHSP70 in Chlamydomonas. Mar. Biotechnol 2012, 14, 332–342. [Google Scholar]
- Sung, D.Y.; Guy, C.L. Physiological and molecular assessment of altered expression of Hsc70-1 in Arabidopsis. Evidence for pleiotropic consequences. Plant Physiol 2003, 132, 979–987. [Google Scholar]
- Katiyar-Agarwal, S.; Agarwal, M.; Grover, A. Heat-tolerant basmati rice engineered by over-expression of hsp101. Plant Mol. Biol 2003, 51, 677–686. [Google Scholar]
- Neilson, K.A.; Gammulla, G.; Mirzaei, M.; Imin, N.; Haynes, P.A. Proteomic analysis of temperature stress in plants. Proteomics 2010, 10, 828–845. [Google Scholar]
- Yamada, A.; Sekiguchi, M.; Mimura, T.; Ozeki, Y. The role of plant CCTα in salt- and osmotic-stress tolerance. Plant Cell Physiol 2002, 43, 1043–1048. [Google Scholar]
- Goswami, A.; Banerjee, R.; Raha, S. Mechanisms of plant adaptation/memory in rice seedlings under arsenic and heat stress: Expression of heat-shock protein gene HSP70. AoB Plants 2010, 2010. [Google Scholar] [CrossRef]
- Cao, F.; Cheng, H.; Cheng, S.; Li, L.; Xu, F.; Yu, W.; Yuan, H. Expression of selected ginkgo biloba heat shock protein genes after cold treatment could be induced by other abiotic stress. Int. J. Mol. Sci 2012, 13, 5768–5788. [Google Scholar]
- Song, H.M.; Wang, H.Z.; Xu, X.B. Overexpression of AtHsp90.3 in Arabidopsis thaliana impairs plant tolerance to heavy metal stress. Biol. Plant 2012, 56, 197–199. [Google Scholar]
- Grigorova, B.; Vaseva, I.I.; Demirevska, K.; Feller, U. Expression of selected heat shock proteins after individually applied and combined drought and heat stress. Acta Physiol. Plant 2011, 33, 2041–2049. [Google Scholar]
- Munns, R. Comparative physiology of salt and water stress. Plant Cell Environ 2002, 25, 239–250. [Google Scholar]
- Sahi, C.; Singh, A.; Blumwald, E.; Grover, A. Beyond osmolytes and transporters: Novel plant salt-stress tolerance-related genes from transcriptional profiling data. Physiol. Plant 2006, 127, 1–9. [Google Scholar]
- Ashraf, M.; Akram, N.A. Improving salinity tolerance of plants through conventional breeding and genetic engineering: An analytical comparison. Biotechnol. Adv 2009, 27, 744–752. [Google Scholar]
- Ji, X.; Wang, Y.; Liu, G. Expression analysis of MYC genes from Tamarix hispida in response to different abiotic stresses. Int. J. Mol. Sci 2012, 13, 1300–1313. [Google Scholar]
- Yang, O.; Popova, O.V.; Süthoff, U.; Lüking, I.; Dietz, K.-J.; Golldac, D. The Arabidopsis basic leucine zipper transcription factor AtbZIP24 regulates complex transcriptional networks involved in abiotic stress resistance. Gene 2009, 436, 45–55. [Google Scholar]
- Costa, C.N.M.; Santa-Brígida, A.B.; Borges, B.N.; Menezes-Neto, M.A.; Carvalho, L.J.C.B.; de Souza, C.R.B. Levels of MeLEA3, a cDNA sequence coding for an atypical late embryogenesis abundant protein in cassava, increase under in vitro salt stress treatment. Plant Mol. Biol. Rep 2011, 29, 997–1005. [Google Scholar]
- Reis, S.P.; Tavares, L.S.C.; Costa, C.N.M.; Santa-Brígida, A.B.; de Souza, C.R.B. Molecular cloning and characterization of a novel RING zinc-finger protein gene up-regulated under in vitro salt stress in cassava. Mol. Biol. Rep 2012, 39, 6513–6519. [Google Scholar]
- Kishor, P.B.K.; Sangam, S.; Amrutha, R.N.; Laxmi, P.S.; Naidu, K.R.; Rao, K.R.S.S.; Rao, S.; Reddy, K.J.; Theriappan, P.; Sreenivasulu, N. Regulation of proline biosynthesis, degradation, uptake and transport in higher plants: Its implications in plant growth and abiotic stress tolerance. Curr. Sci 2005, 88, 424–438. [Google Scholar]
- Jha, Y.; Subramanian, R.B.; Patel, S. Combination of endophytic and rhizospheric plant growth promoting rhizobacteria in Oryza sativa shows higher accumulation of osmoprotectant against saline stress. Acta Physiol. Plant 2011, 33, 797–802. [Google Scholar]
- Ashraf, M.; Foolad, M.R. Roles of glycine betaine and proline in improving plant abiotic stress resistance. Environ. Exp. Bot 2007, 59, 206–216. [Google Scholar]
- Verbruggen, N.; Hermans, C. Proline accumulation in plants: A review. Amino Acids 2008, 35, 753–759. [Google Scholar]
- Stein, H.; Honig, A.; Miller, G.; Erster, O.; Eilenberg, H.; Csonka, L.N.; Szabados, L.; Koncz, C.; Zilberstein, A. Elevation of free proline and proline-rich protein levels by simultaneous manipulations of proline biosynthesis and degradation in plants. Plant Sci 2011, 181, 140–150. [Google Scholar]
- Chaitanya, K.V.; Rasineni, G.K.; Reddy, A.R. Biochemical responses to drought stress in mulberry (Morus alba L.): Evaluation of proline, glycine betaine and abscisic acid accumulation in five cultivars. Acta Physiol. Plant 2009, 31, 437–443. [Google Scholar]
- Silva-Ortega, C.O.; Ochoa-Alfaro, A.E.; Reyes-Agüero, J.A.; Aguado-Santacruz, G.A.; Jiménez-Bremont, J.F. Salt stress increases the expression of p5cs gene and induces proline accumulation in cactus pear. Plant Physiol. Biochem 2008, 46, 82–92. [Google Scholar]
- Székely, G.; Ábrahám, E.; Cséplo, A.; Rigó, G.; Zsigmond, L.; Csiszár, J.; Ayaydin, F.; Strizhov, N.; Jásik, J.; Schmelzer, E.; et al. Duplicated P5CS genes of Arabidopsis play distinct roles in stress regulation and developmental control of proline biosynthesis. Plant J 2008, 53, 11–28. [Google Scholar]
- Rocha, I.M.A.; Vitorello, V.A.; Silva, J.S.; Ferreira-Silva, S.L.; Viégas, R.A.; Silva, E.N.; Silveira, J.A.G. Exogenous ornithine is an effective precursor and the δ-ornithine amino transferase pathway contributes to proline accumulation under high N recycling in salt-stressed cashew leaves. J. Plant Physiol 2012, 169, 41–49. [Google Scholar]
- Yang, G.; Zhou, R.; Tang, T.; Chen, X.; Ouyang, J.; He, L.; Li, W.; Chen, S.; Guo, M.; Li, X.; Zhong, C.; Shi, S. Gene Expression Profiles in Response to Salt Stress in Hibiscus tiliaceus. Plant. Mol. Biol. Rep 2011, 29, 609–617. [Google Scholar]
- Chen, T.H.H.; Murata, N. Glycinebetaine protects plants against abiotic stress: Mechanisms and biotechnological applications. Plant Cell Environ 2011, 34, 1–20. [Google Scholar]
- Chen, T.H.H.; Murata, N. Glycinebetaine: An effective protectant against abiotic stress in plants. Trends Plant Sci 2008, 139, 499–505. [Google Scholar]
- Peleg, Z.; Apse, M.P.; Blumwald, E. Engineering salinity and water-stress tolerance in crop plants: Getting closer to the field. Adv. Bot. Res 2011, 57, 405–443. [Google Scholar]
- Khan, M.S.; Yu, X.; Kikuchi, A.; Asahina, M.; Watanabe, K.N. Genetic engineering of glycine betaine biosynthesis to enhance abiotic stress tolerance in plants. Plant Biotechnol 2009, 26, 125–134. [Google Scholar]
- Banu, M.N.A.; Hoque, M.A.; Watanabe-Sugimoto, M.; Matsuoka, K.; Nakamura, Y.; Shimoishi, Y.; Murata, Y. Proline and glycinebetaine induce antioxidant defense gene expression and suppress cell death in cultured tobacco cells under salt stress. J. Plant Physiol 2009, 166, 146–156. [Google Scholar]
- Chen, S.; Gollop, N.; Heuer, B. Proteomic analysis of salt-stressed tomato (Solanum lycopersicum) seedlings: Effect of genotype and exogenous application of glycinebetaine. J. Exp. Bot 2009, 607, 2005–2019. [Google Scholar]
- Hassine, A.B.; Ghanem, M.E.; Bouzid, S.; Lutts, S. An inland and a coastal population of the Mediterranean xero-halophyte species Atriplex halimus L. differ in their ability to accumulate proline and glycinebetaine in response to salinity and water stress. J. Exp. Bot 2008, 596, 1315–1326. [Google Scholar]
- Mahouachi, J.; Argamasilla, R.; Gómez-Cadenas, A. Influence of exogenous glycine betaine and abscisic acid on papaya in responses to water-deficit stress. J. Plant Growth Regul 2012, 31, 1–10. [Google Scholar]
- Nawaz, K.; Ashraf, M. Exogenous application of glycinebetaine modulates activities of antioxidants in maize plants subjected to salt stress. J. Agron. Crop Sci 2010, 196, 28–37. [Google Scholar]
- Rezaei, M.A.; Kaviani, B.; Jahanshahi, H. Application of exogenous glycine betaine on some growth traits of soybean (Glycine max L.) cv. DPX in drought stress conditions. Sci. Res. Essays 2012, 7, 432–436. [Google Scholar]
- Islam, M.M.; Hoque, M.A.; Okuma, E.; Banu, M.N.A.; Shimoishi, Y.; Nakamura, Y.; Murata, Y. Exogenous proline and glycinebetaine increase antioxidant enzyme activities and confer tolerance to cadmium stress in cultured tobacco cells. J. Plant Physiol 2009, 166, 1587–1597. [Google Scholar]
- Li, S.; Li, F.; Wang, J.; Zhang, W.; Meng, Q.; Chen, T.H.H.; Murata, N.; Yang, X. Glycinebetaine enhances the tolerance of tomato plants to high temperature during germination of seeds and growth of seedlings. Plant Cell Environ 2011, 34, 1931–1943. [Google Scholar]
- Iqbal, N.; Ashraf, Y.; Ashraf, M. Modulation of endogenous levels of some key organic metabolites by exogenous application of glycine betaine in drought stressed plants of sunflower (Helianthus annuus L.). Plant Growth Regul 2011, 63, 7–12. [Google Scholar]
- Duan, X.G.; Song, Y.J.; Yang, A.F.; Zhang, J.R. The transgene pyramiding tobacco with betaine synthesis and heterologous expression of AtNHX1 is more tolerant to salt stress than either of the tobacco lines with betaine synthesis or AtNHX1. Physiol. Plant 2009, 135, 281–295. [Google Scholar]
- He, C.; Yang, A.; Zhang, W.; Gao, Q.; Zhang, J. Improved salt tolerance of transgenic wheat by introducing beta gene for glycine betaine synthesis. Plant Cell Tissue Organ Cult 2010, 101, 65–78. [Google Scholar]
- He, C.; Zhang, W.; Gao, Q.; Yang, A.; Hu, X.; Zhang, J. Enhancement of drought resistance and biomass by increasing the amount of glycine betaine in wheat seedlings. Euphytica 2011, 177, 151–167. [Google Scholar]
- Zhang, K.; Guo, N.; Lian, L.; Wang, J.; Lv, S.; Zhang, J. Improved salt tolerance and seed cotton yield in cotton (Gossypium hirsutum L.) by transformation with betA gene for glycinebetaine synthesis. Euphytica 2011, 181, 1–16. [Google Scholar]
- Yang, X.; Liang, Z.; Wen, X.; Lu, C. Genetic engineering of the biosynthesis of glycinebetaine leads to increased tolerance of photosynthesis to salt stress in transgenic tobacco plants. Plant Mol. Biol 2008, 66, 73–86. [Google Scholar]
- Zhou, S.; Chen, X.; Zhang, X.; Li, Y. Improved salt tolerance in tobacco plants by co-transformation of a betaine synthesis gene BADH and a vacuolar Na+/H+ antiporter gene SeNHX1. Biotechnol. Lett 2008, 30, 369–376. [Google Scholar]
- Wang, G.P.; Zhang, X.Y.; Li, F.; Luo, Y.; Wang, W. Overaccumulation of glycine betaine enhances tolerance to drought and heat stress in wheat leaves in the protection of photosynthesis. Photosynthetica 2010, 48, 117–126. [Google Scholar]
- Zhang, X.Y.; Liang, C.; Wang, G.P.; Luo, Y.; Wang, W. The protection of wheat plasma membrane under cold stress by glycine betaine overproduction. Biol. Plant 2010, 54, 83–88. [Google Scholar]
- Kathuria, H.; Giri, J.; Nataraja, K.N.; Murata, N.; Udayakumar, M.; Tyagi, A.K. Glycinebetaine-induced water-stress tolerance in codA-expressing transgenic indica rice is associated with up-regulation of several stress responsive genes. Plant Biotechnol. J 2009, 7, 512–526. [Google Scholar]
- Luo, Y.; Li, W.-M.; Wang, W. Trehalose: Protector of antioxidant enzymes or reactive oxygen species scavenger under heat stress? Environ. Exp Bot 2008, 63, 378–384. [Google Scholar]
- Schluepmann, H.; Berke, L.; Sanchez-Perez, G.F. Metabolism control over growth: A case for trehalose-6-phosphate in plants. J. Exp. Bot 2011. [Google Scholar] [CrossRef]
- Li, H.-W.; Zang, B.-S.; Deng, X.-W.; Wang, X.-P. Overexpression of the trehalose-6-phosphate synthase gene OsTPS1 enhances abiotic stress tolerance in rice. Planta 2011, 234, 1007–1018. [Google Scholar]
- Fernandez, O.; Béthencourt, L.; Quero, A.; Sangwan, R.S.; Clément, C. Trehalose and plant stress responses: Friend or foe? Trends Plant Sci 2010, 15, 409–417. [Google Scholar]
- Ge, L.-F.; Chao, D.-Y.; Shi, M.; Zhu, M.-Z.; Gao, J.-P.; Lin, H.-X. Overexpression of the trehalose-6-phosphate phosphatase gene OsTPP1 confers stress tolerance in rice and results in the activation of stress responsive genes. Planta 2008, 228, 191–201. [Google Scholar]
- Redillas, M.C.F.R.; Park, S.-H.; Lee, J.W.; Kim, Y.S.; Jeong, J.S.; Jung, H.; Bang, S.W.; Hahn, T.R.; Kim, J.-K. Accumulation of trehalose increases soluble sugar contents in rice plants conferring tolerance to drought and salt stress. Plant. Biotechnol. Rep 2012, 6, 89–96. [Google Scholar]
- Rodríguez-Salazar, J.; Suárez, R.; Caballero-Mellado, J.; Iturriaga, G. Trehalose accumulation in Azospirillum brasilense improves drought tolerance and biomass in maize plants. FEMS Microbiol. Lett 2009, 296, 52–59. [Google Scholar]
- Stiller, I.; Dulai, S.; Kondrák, M.; Tarnai, R.; Szabó, L.; Toldi, O.; Bánfalvi, Z. Effects of drought on water content and photosynthetic parameters in potato plants expressing the trehalose-6-phosphate synthase gene of Saccharomyces cerevisiae. Planta 2008, 227, 299–308. [Google Scholar]
- Suárez, R.; Calderón, C.; Iturriaga, G. Enhanced Tolerance to Multiple Abiotic Stresses in Transgenic Alfalfa Accumulating Trehalose. Crop Sci 2009, 49, 1791–1799. [Google Scholar]
- Sugawara, M.; Cytryn, E.J.; Sadowsky, M.J. Functional role of Bradyrhizobium japonicum trehalose biosynthesis and metabolism genes during physiological stress and nodulation. Appl. Environ. Microbiol 2010, 76, 1071–1081. [Google Scholar]
- Li, H.; Wang, H.-L.; Du, J.; Du, G.; Zhan, J.-C.; Huang, W.-D. Trehalose protects wine yeast against oxidation under thermal stress. World J. Microbiol. Biotechnol 2010, 26, 969–976. [Google Scholar]
- López, M.; Tejera, N.A.; Lluch, C. ValidamycinA improves the response of Medicago truncatula plants to salt stress by inducing trehalose accumulation in the root nodules. J. Plant Physiol 2009, 166, 1218–1222. [Google Scholar]
- Nounjan, N.; Nghia, P.T.; Theerakulpisut, P. Exogenous proline and trehalose promote recovery of rice seedlings from salt-stress and differentially modulate antioxidant enzymes and expression of related genes. J. Plant Physiol 2012, 169, 596–604. [Google Scholar]
- Zeid, I.M. Trehalose as osmoprotectant for maize under salinity-induced stress. Res. J. Agric. Biol. Sci 2009, 5, 613–622. [Google Scholar]
- Gupta, A.K.; Kaur, N. Sugar signalling and gene expression in relation to carbohydrate metabolism under abiotic stresses in plants. J. Biosci 2005, 30, 761–776. [Google Scholar]
- Conde, C.; Silva, P.; Agasse, A.; Lemoine, R.; Delrot, S.; Tavares, R.; Geros, H. Utilization and transport of mannitol in Olea europaea and implications for salt stress tolerance. Plant Cell Physiol 2007, 48, 42–53. [Google Scholar]
- Tang, W.; Peng, X.; Newton, R.J. Enhanced tolerance to salt stress in transgenic loblolly pine simultaneously expressing two genes encoding mannitol-1-phosphatedehydrogenase and glucitol-6-phosphate dehydrogenase. Plant Physiol. Biochem 2005, 43, 139–146. [Google Scholar]
- Abebe, T.; Guenzi, A.C.; Martin, B.; Cushman, J.C. Tolerance of mannitol-accumulating transgenic wheat to water stress and salinity. Plant Physiol 2003, 131, 1748–1755. [Google Scholar]
- Chan, Z.; Grumet, R.; Loescher, W. Global gene expression analysis of transgenic, mannitol producing, and salt-tolerant Arabidopsis thaliana indicates widespread changes in abiotic and biotic stress-related genes. J. Exp. Bot 2011, 62, 4787–4803. [Google Scholar]
- Tari, I.; Kiss, G.; Deér, A.K.; Csiszár, J.; Erdei, L.; Gallé, A.; Gémes, K.; Horváth, F.; Poór, P.; Szepesi, Á.; et al. Salicylic acid increased aldose reductase activity and sorbitol accumulation in tomato plants under salt stress. Biol. Plant 2010, 54, 677–683. [Google Scholar]
- Feng, X.; Zhao, P.; Hao, J.; Hu, J.; Kang, D.; Wang, H. Effects of sorbitol on expression of genes involved in regeneration of upland rice (Oryza sativa L.). Plant Cell Tissue Organ Cult 2011, 106, 455–463. [Google Scholar]
- Li, F.; Lei, H.; Zhao, X.; Tian, R.; Li, T. Characterization of three sorbitol transporter genes in micropropagated apple plants grown under drought stress. Plant Mol. Biol. Rep 2012, 30, 123–130. [Google Scholar]
- Kanamaru, N.; Ito, Y.; Komori, S.; Saito, M.; Kato, H.; Takahashi, S.; Omura, M.; Soejima, J.; Shiratake, K.; Yamada, K.; Yamaki, S. Transgenic apple transformed by sorbitol-6-phosphate dehydrogenase cDNA Switch between sorbitol and sucrose supply due to its gene expression. Plant Sci 2004, 167, 55–61. [Google Scholar]
- Liang, D.; Cui, M.; Wu, S.; Ma, F.-W. Genomic structure, sub-cellular localization, and promoter analysis of the gene encoding sorbitol-6–phosphate dehydrogenase from apple. Plant Mol. Biol. Rep 2012, 30, 904–914. [Google Scholar]
- Gao, Z.; Jayanty, S.; Beaudry, R.; Loescher, W. Sorbitol transporter expression in apple sink tissues: Implications for fruit sugar accumulation and watercore development. J. Am. Soc. Hort. Sci 2005, 130, 261–268. [Google Scholar]
- Fan, R.-C.; Peng, C.-C.; Xu, Y.-H.; Wang, X.-F.; Li, Y.; Shang, Y.; Du, S.-Y.; Zhao, R.; Zhang, X.-Y.; Zhang, L.-Y.; et al. Apple sucrose transporter SUT1 and sorbitol transporter SOT6 interact with Cytochrome b5 to regulate their affinity for substrate sugars. Plant Physiol 2009, 150, 1880–1901. [Google Scholar]
- Wormit, A.; Trentmann, O.; Feifer, I.; Lohr, C.; Tjaden, J.; Meyer, S.; Schmidt, U.; Martinoia, E.; Neuhaus, H.E. molecular identification and physiological characterization of a novel monosaccharide transporter from Arabidopsis involved in vacuolar sugar transport. Plant Cell 2006, 18, 3476–3490. [Google Scholar]
- Pommerrenig, B.; Papini-Terzi, F.S.; Sauer, N. Differential regulation of sorbitol and sucrose loading into the phloem of Plantago major in response to salt stress. Plant Physiol 2007, 144, 1029–1038. [Google Scholar]
- Dure, L., III; Greenway, S.C.; Galau, G.A. Developmental biochemistry of cottonseed embryogenesis and germination: Changing messenger ribonucleic acid populations as shown by in vitro and in vivo protein synthesis. Biochemistry 1981, 20, 4162–4168. [Google Scholar]
- Bray, E.A. Molecular responses to water deficit. Plant Physiol 1993, 103, 1035–1040. [Google Scholar]
- Integram, J.; Bartels, D. The molecular basis of dehydration tolerance in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol 1996, 47, 377–403. [Google Scholar]
- Bray, E.A. Plant responses to water deficit. Trends Plant Sci 1997, 2, 48–54. [Google Scholar]
- Wang, W.; Vinocur, B.; Altman, A. Plant responses to drought, salinity, and extreme temperatures: Towards genetic engineering for stress tolerance. Planta 2003, 218, 1–14. [Google Scholar]
- Tunnacliffe, A.; Wise, M.J. The continuing conundrum of the LEA proteins. Naturwissenschaften 2007, 94, 791–812. [Google Scholar]
- Battaglia, M.; Olvera-Carrillo, Y.; Garciarrubio, A.; Campos, F.; Covarrubias, A.A. The enigmatic LEA proteins and other hydrophilins. Plant Physiol 2008, 148, 6–24. [Google Scholar]
- Shih, M.D.; Hoekstra, F.A.; Hsing, Y.I.C. Late embryogenesis abundant proteins. Adv. Bot. Res 2008, 48, 211–255. [Google Scholar]
- Dure, L., III; Crouch, M.; Harada, J.; Ho, T.H.D.; Mundy, J.; Quatrano, R.S.; Thomas, T.; Sung, Z.R. Common amino acid sequence domains among the LEA proteins of higher plants. Plant Mol. Biol 1989, 12, 475–486. [Google Scholar]
- Hand, C.S.; Menze, M.A.; Toner, M.; Boswell, L.; Moore, D. LEA proteins during water stress: Not just for plants anymore. Annu. Rev. Physiol 2011, 73, 115–134. [Google Scholar]
- Browne, J.; Tunnacliffe, A.; Burnell, A. Anhydrobiosis: Plant desiccation gene found in a nematode. Nature 2002, 416. [Google Scholar] [CrossRef]
- Browne, J.A.; Dolan, K.M.; Tyson, T.; Goyal, K.; Tunnacliffe, A.; Burnell, A.M. Dehydration-specific induction of hydrophilic protein genes in the anhydrobiotic nematode Aphelenchus avenae. Eukaryot. Cell 2004, 3, 966–975. [Google Scholar]
- Gal, T.Z.; Glazer, I.; Koltai, H. Differential gene expression during desiccation stress in the insect-killing nematode Steinernema feltlae IS-6. J. Parasitol 2003, 89, 761–766. [Google Scholar]
- Gal, T.Z.; Glazer, I.; Koltai, H. An LEA group 3 family member is involved in survival of C. elegans during exposure to stress. FEBS Lett 2004, 577, 21–26. [Google Scholar]
- Goyal, K.; Pinelli, C.; Maslen, S.L.; Rastogi, R.K.; Stephens, E.; Tunnacliffe, A. Dehydration-regulated processing of late embryogenesis abundant protein in a desiccationtolerant nematode. FEBS Lett 2005, 579, 4093–4098. [Google Scholar]
- He, J.X.; Fu, J.R. Research progress in Lea proteins of seeds. Plant Physiol. Commun 1996, 32, 241–246. [Google Scholar]
- Wise, M.J. LEAping to conclusions: A computational reanalysis of late embryogenesis abundant proteins and their possible roles. BMC Bioinform 2003, 4, 1–19. [Google Scholar]
- Swire-Clark, G.A.; Marcotte, W.R. The wheat LEA protein Em functions as an osmoprotective molecule in Saccharomyces cerevisiae. Plant Mol. Biol 1999, 39, 117–128. [Google Scholar]
- Xiao, B.; Huang, Y.; Tang, N.; Xiong, L. Over-expression of a LEA gene in rice improves drought resistance under the field conditions. Theor. Appl. Genet 2007, 115, 35–46. [Google Scholar]
- Lopez, C.G.; Banowetz, G.M.; Peterson, C.J.; Kronstad, W.E. Dehydrin expression and drought tolerance in seven wheat cultivars. Crop Sci 2003, 43, 577–582. [Google Scholar]
- Babu, R.C.; Zhang, J.; Blum, A.; Ho, T.H.D.; Wu, R.; Nguyen, H.T. HVA1, a LEA gene from barley confers dehydration tolerance in transgenic rice (Oryza sativa L.) via cell membrane protection. Plant Sci 2004, 166, 855–862. [Google Scholar]
- Maqbool, B.; Zhong, H.; El Maghraby, Y.; Ahmad, A.; Chai, B.; Wang, W.; Sabzikar, R.; Sticklen, B. Competence of oat (Avena sativa L.) shoot apical meristems for integrative transformation, inherited expression and osmotic tolerance of transgenic lines containing hva1. Theor. Appl. Genet 2002, 105, 201–208. [Google Scholar]
- Sivamani, E.; Bahieldinl, A.; Wraith, J.M.; Al Niemi, T.; Dyer, W.E.; Ho, T.H.D.; Qu, R. Improved biomass productivity and water use efficiency under water deficit conditions in transgenic wheat constitutively expressing the barley HVA1 gene. Protein Sci 2000, 155, 1–9. [Google Scholar]
- NDong, C.; Danyluk, J.; Wilson, K.E.; Pocock, T.; Huner, N.P.; Sarhan, F. Cold-regulated cereal chloroplast late embryogenesis abundant-like proteins. Molecular characterization and functional analyses. Plant Physiol 2002, 129, 1368–1381. [Google Scholar]
- Borrell, A.; Cutanda, M.C.; Lumbreras, V.; Pujal, J.; Goday, A.; Culiáñez-Macià, F.A.; Pagès, M. Arabidopsis thaliana atrab28: A nuclear targeted protein related to germination and toxic cation tolerance. Plant Mol. Biol 2002, 50, 249–259. [Google Scholar]
- Hara, M.; Terashima, S.; Fukaya, T.; Kuboi, T. Enhancement of cold tolerance and inhibition of lipid peroxidation by citrus dehydrin in transgenic tobacco. Planta 2003, 217, 290–298. [Google Scholar]
- Park, B.J.; Liu, Z.C.; Kanno, A.; Kameya, T. Genetic improvement of Chinese cabbage for salt and drought tolerance by constitutive expression of a B. napus LEA gene. Plant Sci 2005, 169, 553–558. [Google Scholar]
- Park, B.J.; Liu, Z.C.; Kanno, A.; Kameya, T. Increased tolerance to salt and water deficit stress in transgenic lettuce (Lactuca sativa L.) by constitutive expression of LEA. Plant Growth Regul 2005, 45, 165–171. [Google Scholar]
- Figueras, M.; Pujal, J.; Saleh, A.; Save, R.; Pages, M.; Goday, A. Maize Rab17 overexpression in Arabidopsis plants promotes osmotic stress tolerance. Ann. Appl. Biol 2004, 144, 251–257. [Google Scholar]
- Peng, W. GM crop cultivation surges, but novels traits languish. Nat. Biotechnol 2011, 29. [Google Scholar] [CrossRef]
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Dos Reis, S.P.; Lima, A.M.; De Souza, C.R.B. Recent Molecular Advances on Downstream Plant Responses to Abiotic Stress. Int. J. Mol. Sci. 2012, 13, 8628-8647. https://doi.org/10.3390/ijms13078628
Dos Reis SP, Lima AM, De Souza CRB. Recent Molecular Advances on Downstream Plant Responses to Abiotic Stress. International Journal of Molecular Sciences. 2012; 13(7):8628-8647. https://doi.org/10.3390/ijms13078628
Chicago/Turabian StyleDos Reis, Sávio Pinho, Aline Medeiros Lima, and Cláudia Regina Batista De Souza. 2012. "Recent Molecular Advances on Downstream Plant Responses to Abiotic Stress" International Journal of Molecular Sciences 13, no. 7: 8628-8647. https://doi.org/10.3390/ijms13078628
APA StyleDos Reis, S. P., Lima, A. M., & De Souza, C. R. B. (2012). Recent Molecular Advances on Downstream Plant Responses to Abiotic Stress. International Journal of Molecular Sciences, 13(7), 8628-8647. https://doi.org/10.3390/ijms13078628