Secretome Survey of Human Plexiform Neurofibroma Derived Schwann Cells Reveals a Secreted form of the RARRES1 Protein
Abstract
:1. Introduction
2. Results and Discussion
2.1. Primary Schwann Cell Isolation and Culture
2.2. Secretome Profiling
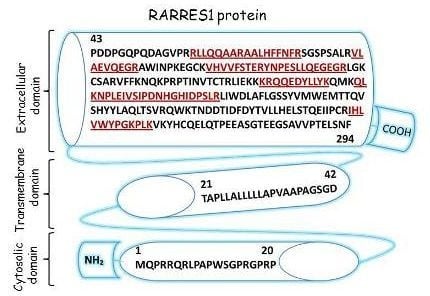
2.3. Characterization of Schwann Cell Secreted RARRES1 Protein
2.4. mRNA RARRES Expression Levels in NF1 Versus Normal Schwann Cells
2.4.1. Quantitative RTPCR
2.4.2. Western Blot Analysis
2.5. All-Trans Retinoic Acid Induces Expression of Secreted RARRES1
2.6. Discussion
3. Experimental Section
3.1. Specimen Collection
3.2. Isolation and Culture of Human Primary Schwann Cells
3.3. Secretome Profiling
3.4. Liquid Chromatography-Tandem Mass Spectrometry
3.5. Database Search and Quantification
3.6. Reverse-Transcription PCR
3.7. Western Blot Analysis
3.8. Effect of All-Trans Retinoic Acid on the sNF96.2 Secreted RARRES1
4. Conclusions
Acknowledgments
References
- Goldberg, N.S.; Collins, F.S. The hunt for the neurofibromatosis gene. Arch. Dermatol 1991, 127, 1705–1707. [Google Scholar]
- Korf, B.R. Plexiform neurofibromas. Am. J. Med. Genet 1999, 89, 31–37. [Google Scholar]
- Parrinello, S.; Lloyd, A.C. Neurofibroma development in NF1--insights into tumour initiation. Trends Cell Biol 2009, 19, 395–403. [Google Scholar]
- Lantieri, L.; Meningaud, J.P.; Grimbert, P.; Bellivier, F.; Lefaucheur, J.P.; Ortonne, N.; Benjoar, M.D.; Lang, P.; Wolkenstein, P. Repair of the lower and middle parts of the face by composite tissue allotransplantation in a patient with massive plexiform neurofibroma: A 1-year follow-up study. Lancet 2008, 372, 639–645. [Google Scholar]
- Needle, M.N.; Cnaan, A.; Dattilo, J.; Chatten, J.; Phillips, P.C.; Shochat, S.; Sutton, L.N.; Vaughan, S.N.; Zackai, E.H.; Zhao, H.; et al. Prognostic signs in the surgical management of plexiform neurofibroma: The Children’s Hospital of Philadelphia experience, 1974–1994. J. Pediatr 1997, 131, 678–682. [Google Scholar]
- Serletis, D.; Parkin, P.; Bouffet, E.; Shroff, M.; Drake, J.M.; Rutka, J.T. Massive plexiform neurofibromas in childhood: Natural history and management issues. J. Neurosurg 2007, 106, S363–S367. [Google Scholar]
- Lee, M.J.; Stephenson, D.A. Recent developments in neurofibromatosis type 1. Curr. Opin. Neurol 2007, 20, 135–141. [Google Scholar]
- Bhatheja, K.; Field, J. Schwann cells: Origins and role in axonal maintenance and regeneration. Int. J. Biochem. Cell Biol 2006, 38, 1995–1999. [Google Scholar]
- Serra, E.; Rosenbaum, T.; Winner, U.; Aledo, R.; Ars, E.; Estivill, X.; Lenard, H.G.; Lazaro, C. Schwann cells harbor the somatic NF1 mutation in neurofibromas: Evidence of two different Schwann cell subpopulations. Hum. Mol. Genet 2000, 9, 3055–3064. [Google Scholar]
- Johansson, G.; Mahller, Y.Y.; Collins, M.H.; Kim, M.O.; Nobukuni, T.; Perentesis, J.; Cripe, T.P.; Lane, H.A.; Kozma, S.C.; Thomas, G.; et al. Effective in vivo targeting of the mammalian target of rapamycin pathway in malignant peripheral nerve sheath tumors. Mol. Cancer Ther 2008, 7, 1237–1245. [Google Scholar]
- Kissil, J.L.; Blakeley, J.O.; Ferner, R.E.; Huson, S.M.; Kalamarides, M.; Mautner, V.F.; McCormick, F.; Morrison, H.; Packer, R.; Ramesh, V.; et al. What’s new in neurofibromatosis? Proceedings from the 2009 NF Conference: New frontiers. Am. J. Med. Genet. A 2012, 152A, 269–283. [Google Scholar]
- Staser, K.; Yang, F.C.; Clapp, D.W. Plexiform neurofibroma genesis: Questions of Nf1 gene dose and hyperactive mast cells. Curr. Opin. Hematol 2010, 17, 287–293. [Google Scholar]
- Yang, F.C.; Ingram, D.A.; Chen, S.; Zhu, Y.; Yuan, J.; Li, X.; Yang, X.; Knowles, S.; Horn, W.; Li, Y.; et al. Nf1-dependent tumors require a microenvironment containing Nf1+/-- and c-kit-dependent bone marrow. Cell 2008, 135, 437–448. [Google Scholar]
- Perrin, G.Q.; Li, H.; Fishbein, L.; Thomson, S.A.; Hwang, M.S.; Scarborough, M.T.; Yachnis, A.T.; Wallace, M.R.; Mareci, T.H.; Muir, D. An orthotopic xenograft model of intraneural NF1 MPNST suggests a potential association between steroid hormones and tumor cell proliferation. Lab. Invest 2007, 87, 1092–1102. [Google Scholar]
- Nagpal, S.; Patel, S.; Asano, A.T.; Johnson, A.T.; Duvic, M.; Chandraratna, R.A. Tazarotene-induced gene 1 (TIG1), a novel retinoic acid receptor-responsive gene in skin. J. Invest. Dermatol 1996, 106, 269–274. [Google Scholar]
- Sahab, Z.J.; Hall, M.D.; Zhang, L.; Cheema, A.K.; Byers, S.W. Tumor Suppressor RARRES1 Regulates DLG2, PP2A, VCP, EB1, and Ankrd26. J. Cancer 2010, 1, 14–22. [Google Scholar]
- Wakoh, T.; Uekawa, N.; Terauchi, K.; Sugimoto, M.; Ishigami, A.; Shimada, J.; Maruyama, M. Implication of p53-dependent cellular senescence related gene, TARSH in tumor suppression. Biochem. Biophys. Res. Commun 2009, 380, 807–812. [Google Scholar]
- Cheng, T.W.; Gong, Q. Secreted TARSH regulates olfactory mitral cell dendritic complexity. Eur. J. Neurosci 2009, 29, 1083–1095. [Google Scholar]
- Hunter, S.; Weiss, S.; Ou, C.Y.; Jaye, D.; Young, A.; Wilcox, J.; Arbiser, J.L.; Monson, D.; Goldblum, J.; Nolen, J.D.; et al. Apolipoprotein D is down-regulated during malignant transformation of neurofibromas. Hum. Pathol 2005, 36, 987–993. [Google Scholar]
- West, R.B.; Harvell, J.; Linn, S.C.; Liu, C.L.; Prapong, W.; Hernandez-Boussard, T.; Montgomery, K.; Nielsen, T.O.; Rubin, B.P.; Patel, R.; et al. Apo D in soft tissue tumors: A novel marker for dermatofibrosarcoma protuberans. Am. J. Surg. Pathol 2004, 28, 1063–1069. [Google Scholar]
- Abelev, G.I. Production of embryonal serum alpha-globulin by hepatomas: Review of experimental and clinical data. Cancer Res 1968, 28, 1344–1350. [Google Scholar]
- Debruyne, E.N.; Delanghe, J.R. Diagnosing and monitoring hepatocellular carcinoma with alpha-fetoprotein: New aspects and applications. Clin. Chim. Acta 2008, 395, 19–26. [Google Scholar]
- Masopust, J.; Kithier, K.; Radl, J.; Koutecky, J.; Kotal, L. Occurrence of fetoprotein in patients with neoplasms and non-neoplastic diseases. Int. J. Cancer 1968, 3, 364–373. [Google Scholar]
- McIntire, K.R.; Waldmann, T.A.; Moertel, C.G.; Go, V.L. Serum alpha-fetoprotein in patients with neoplasms of the gastrointestinal tract. Cancer Res 1975, 35, 991–996. [Google Scholar]
- Li, M.S.; Li, P.F.; Yang, F.Y.; He, S.P.; Du, G.G.; Li, G. The intracellular mechanism of alpha-fetoprotein promoting the proliferation of NIH 3T3 cells. Cell Res 2002, 12, 151–156. [Google Scholar]
- Rao, J.S. Molecular mechanisms of glioma invasiveness: The role of proteases. Nat. Rev. Cancer 2003, 3, 489–501. [Google Scholar]
- Siren, V.; Peltonen, J.; Vaheri, A. Plasminogen activators and their inhibitor gene expression in cutaneous NF1-related neurofibromas. Arch. Dermatol. Res 2006, 297, 421–424. [Google Scholar]
- Aviezer, D.; Hecht, D.; Safran, M.; Eisinger, M.; David, G.; Yayon, A. Perlecan, basal lamina proteoglycan, promotes basic fibroblast growth factor-receptor binding, mitogenesis, and angiogenesis. Cell 1994, 79, 1005–1013. [Google Scholar]
- Camaj, P.; Seeliger, H.; Ischenko, I.; Krebs, S.; Blum, H.; de Toni, E.N.; Faktorova, D.; Jauch, K.W.; Bruns, C.J. EFEMP1 binds the EGF receptor and activates MAPK and Akt pathways in pancreatic carcinoma cells. Biol. Chem 2009, 390, 1293–1302. [Google Scholar]
- Xu, Y.; Tan, L.J.; Grachtchouk, V.; Voorhees, J.J.; Fisher, G.J. Receptor-type protein-tyrosine phosphatase-kappa regulates epidermal growth factor receptor function. J. Biol. Chem 2005, 280, 42694–42700. [Google Scholar]
- Tarcic, G.; Boguslavsky, S.K.; Wakim, J.; Kiuchi, T.; Liu, A.; Reinitz, F.; Nathanson, D.; Takahashi, T.; Mischel, P.S.; Ng, T.; et al. An unbiased screen identifies DEP-1 tumor suppressor as a phosphatase controlling EGFR endocytosis. Curr. Biol 2009, 19, 1788–1798. [Google Scholar]
- Caccia, D.; Zanetti Domingues, L.; Micciche, F.; de Bortoli, M.; Carniti, C.; Mondellini, P.; Bongarzone, I. Secretome compartment is a valuable source of biomarkers for cancer-relevant pathways. J. Proteome Res 2011, 10, 4196–4207. [Google Scholar]
- Wu, C.C.; Hsu, C.W.; Chen, C.D.; Yu, C.J.; Chang, K.P.; Tai, D.I.; Liu, H.P.; Su, W.H.; Chang, Y.S.; Yu, J.S. Candidate serological biomarkers for cancer identified from the secretomes of 23 cancer cell lines and the human protein atlas. Mol. Cell Proteomics 2010, 9, 1100–1117. [Google Scholar]
- An, E.; Gordish-Dressman, H.; Hathout, Y. Effect of TNF-alpha on human ARPE-19-secreted proteins. Mol. Vis 2008, 14, 2292–2303. [Google Scholar]
- An, E.; Sen, S.; Park, S.K.; Gordish-Dressman, H.; Hathout, Y. Identification of novel substrates for the serine protease HTRA1 in the human RPE secretome. Invest. Ophthalmol. Vis. Sci 2010, 51, 3379–3386. [Google Scholar]
- Formolo, C.A.; Williams, R.; Gordish-Dressman, H.; MacDonald, T.J.; Lee, N.H.; Hathout, Y. Secretome signature of invasive glioblastoma multiforme. J. Proteome Res 2011, 10, 3149–3159. [Google Scholar]
- Norheim, F.; Raastad, T.; Thiede, B.; Rustan, A.C.; Drevon, C.A.; Haugen, F. Proteomic identification of secreted proteins from human skeletal muscle cells and expression in response to strength training. Am. J. Physiol. Endocrinol. Metab 2011, 301, E1013–E1021. [Google Scholar]
- Roelofsen, H.; Dijkstra, M.; Weening, D.; de Vries, M.P.; Hoek, A.; Vonk, R.J. Comparison of isotope-labeled amino acid incorporation rates (CILAIR) provides a quantitative method to study tissue secretomes. Mol. Cell Proteomics 2009, 8, 316–324. [Google Scholar]
- Kishi, K.; Imanishi, N.; Ohara, H.; Ninomiya, R.; Okabe, K.; Hattori, N.; Kubota, Y.; Nakajima, H.; Nakajima, T. Distribution of adipose-derived stem cells in adipose tissues from human cadavers. J. Plast. Reconstr. Aesthetic Surg 2010, 63, 1717–1722. [Google Scholar]
- Jing, C.; El-Ghany, M.A.; Beesley, C.; Foster, C.S.; Rudland, P.S.; Smith, P.; Ke, Y. Tazarotene-induced gene 1 (TIG1) expression in prostate carcinomas and its relationship to tumorigenicity. J. Natl. Cancer Inst 2002, 94, 482–490. [Google Scholar]
- Youssef, E.M.; Chen, X.Q.; Higuchi, E.; Kondo, Y.; Garcia-Manero, G.; Lotan, R.; Issa, J.P. Hypermethylation and silencing of the putative tumor suppressor Tazarotene-induced gene 1 in human cancers. Cancer Res 2004, 64, 2411–2417. [Google Scholar]
- Kwok, W.K.; Pang, J.C.; Lo, K.W.; Ng, H.K. Role of the RARRES1 gene in nasopharyngeal carcinoma. Cancer Genet. Cytogenet 2009, 194, 58–64. [Google Scholar]
- Wu, C.C.; Shyu, R.Y.; Chou, J.M.; Jao, S.W.; Chao, P.C.; Kang, J.C.; Wu, S.T.; Huang, S.L.; Jiang, S.Y. RARRES1 expression is significantly related to tumour differentiation and staging in colorectal adenocarcinoma. Eur. J. Cancer 2006, 42, 557–565. [Google Scholar]
- Sahab, Z.J.; Hall, M.D.; Me Sung, Y.; Dakshanamurthy, S.; Ji, Y.; Kumar, D.; Byers, S.W. Tumor suppressor RARRES1 interacts with cytoplasmic carboxypeptidase AGBL2 to regulate the alpha-tubulin tyrosination cycle. Cancer Res 2011, 71, 1219–1228. [Google Scholar]
- Link, A.J.; Eng, J.; Schieltz, D.M.; Carmack, E.; Mize, G.J.; Morris, D.R.; Garvik, B.M.; Yates, J.R., 3rd. Direct analysis of protein complexes using mass spectrometry. Nat. Biotechnol 1999, 17, 676–682. [Google Scholar]





| Donor’s id | Age (years) | Gender | Tumor location | Diagnosis |
|---|---|---|---|---|
| NFSC 141 | 20 | Female | Neck | Plexiform neurofibroma |
| NFSC 142 | 19 | Male | Neck | Plexiform neurofibroma |
| NFSC 143R | 17 | Female | Right lumbar | Plexiform neurofibroma |
| NFSC 143L | Left lumbar | Plexiform neurofibroma |
| Accession n° | Protein name | NSC | NFSCs | Subcellular localization | Function |
|---|---|---|---|---|---|
| P49788 | Retinoic acid receptor responder protein 1 | 0 | 45 | Single-pass membrane | Tumor suppressor |
| P01024 | Complement C3 | 0 | 43 * | secreted | complement pathway |
| P0C0L4 | Complement C4-A | 0 | 39 | secreted | Complement pathway |
| P05090 | Apolipoprotein D | 0 | 15 | secreted | Transporter |
| P22352 | Glutathione peroxidase 3 | 0 | 10 * | secreted | Antioxidant |
| P36222 | Chitinase-3-like protein 1 | 0 | 11 * | secreted | unknown |
| P55268 | Laminin subunit beta-2 | 0 | 17 | secreted | Cell attachment |
| P02649 | Apolipoprotein E | 0 | 7 * | secreted | Transporter |
| Q4LDE5 | Sushi, von Willebrand factor type A | 0 | 5 | membrane/secreted | Cell attachment |
| Q92954 | Proteoglycan 4 | 0 | 5 | secreted | Adhesion inhibition |
| Q12805 | EFEMP1 | 0 | 9 * | secreted | Cell adhesion/migration |
| P02771 | Alpha-fetoprotein | 0 | 6 | secreted | Transporter |
| P55285 | Cadherin-6 | 16 | 0 | Membrane/cell surface | Cell adhesion |
| P24844 | Myosin regulatory light polypeptide 9 | 12 | 0 | cytoskeleton | Cytokenesis and cell locomotion |
| Q13308 | Tyrosine-protein kinase-like 7 | 11 | 0 | cell surface | Wnt signaling pathway |
| P09104 | Gamma-enolase | 9 | 0 | cytosol/membrane | glycogenesis |
| Accession n° | Protein name | NSC | NFSCs | Subcellular localization | Function |
|---|---|---|---|---|---|
| Q16363 | Laminin subunit alpha-4 | 13 | 81 | secreted | cell attachment/migration |
| Q14112 | Nidogen-2 | 25 | 81 | secreted | cell attachment/migration |
| P05155 | Plasma protease C1 inhibitor | 7 | 85 | secreted | C1 complex inhibitor |
| P07585 | Decorin | 12 | 67 | secreted | extracellular remodeling |
| Q7Z7G0 | Target of Nesh-SH3 | 1 | 19 | secreted | cell proliferation |
| P98160 | Perlecan | 5 | 46 | secreted | extracellular matrix remodeling |
| P24592 | Insulin-like growth factor-binding protein 6 | 4 | 16 | secreted | regulates cell growth |
| P05121 | Plasminogen activator inhibitor 1 | 727 | 284 | secreted | inhibitor of fibrinolysis |
| P13611 | Versican core protein | 12 | 1 | secreted | intercellular signaling and connecting cells with the extracellular matrix |
| Q6UVK1 | Chondroitin sulfate proteoglycan 4 | 83 | 19 | Cell surface | cell growth and migration |
| Q15262 | Receptor-type tyrosine-protein phosphatase kappa | 12 | 2 | Cell surface | Negative regulator of EGFR signaling pathway |
| P05556 | Integrin beta-1 | 13 | 1 | Cell surface | Cell adhesion |
© 2012 by the authors; licensee Molecular Diversity Preservation International, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Chen, H.-L.; Seol, H.; Brown, K.J.; Gordish-Dressman, H.; Hill, A.; Gallo, V.; Packer, R.; Hathout, Y. Secretome Survey of Human Plexiform Neurofibroma Derived Schwann Cells Reveals a Secreted form of the RARRES1 Protein. Int. J. Mol. Sci. 2012, 13, 9380-9399. https://doi.org/10.3390/ijms13079380
Chen H-L, Seol H, Brown KJ, Gordish-Dressman H, Hill A, Gallo V, Packer R, Hathout Y. Secretome Survey of Human Plexiform Neurofibroma Derived Schwann Cells Reveals a Secreted form of the RARRES1 Protein. International Journal of Molecular Sciences. 2012; 13(7):9380-9399. https://doi.org/10.3390/ijms13079380
Chicago/Turabian StyleChen, Hui-Ling, Haeri Seol, Kristy Jean Brown, Heather Gordish-Dressman, Ashley Hill, Vittorio Gallo, Roger Packer, and Yetrib Hathout. 2012. "Secretome Survey of Human Plexiform Neurofibroma Derived Schwann Cells Reveals a Secreted form of the RARRES1 Protein" International Journal of Molecular Sciences 13, no. 7: 9380-9399. https://doi.org/10.3390/ijms13079380
APA StyleChen, H. -L., Seol, H., Brown, K. J., Gordish-Dressman, H., Hill, A., Gallo, V., Packer, R., & Hathout, Y. (2012). Secretome Survey of Human Plexiform Neurofibroma Derived Schwann Cells Reveals a Secreted form of the RARRES1 Protein. International Journal of Molecular Sciences, 13(7), 9380-9399. https://doi.org/10.3390/ijms13079380