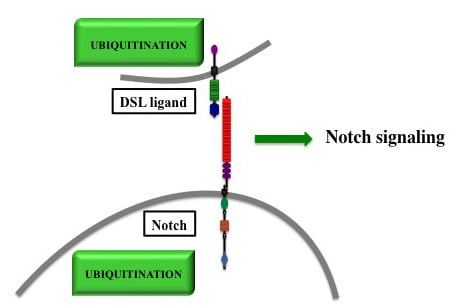
Ubiquitinations in the Notch Signaling Pathway
Abstract
:1. Introduction
2. Main Features of Notch Signal Transduction
3. Ubiquitination, Deubiquitination
4. How Ubiquitinations Regulate Notch Pathway
4.1. NIC Stability
4.2. Notch at the Cell Surface
4.3. Notch Activation Process
4.4. Ligand Maintenance and Signaling Activity
4.4.1. Transendocytosis
4.4.2. Recycling of the Ligands
5. Conclusions
Acknowledgments
Conflict of Interest
References
- Artavanis-Tsakonas, S. Molecular cloning of Notch, a locus affecting neurogenesis in Drosophila melanogaster. Proc. Natl. Acad. Sci. USA 1983, 80, 1977–1981. [Google Scholar]
- Joutel, A.; Corpechot, C.; Ducros, A.; Vahedi, K.; Chabriat, H.; Mouton, P.; Alamowitch, S.; Domenga, V.; Cecillion, M.; Marechal, E.; et al. Notch3 mutations in cadasil, a hereditary adult-onset condition causing stroke and dementia. Nature 1996, 383, 707–710. [Google Scholar]
- Agrawal, N.; Frederick, M.J.; Pickering, C.R.; Bettegowda, C.; Chang, K.; Li, R.J.; Fakhry, C.; Xie, T.-X.; Zhang, J.; Wang, J.; et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in notch1. Science 2011, 333, 1154–1157. [Google Scholar]
- Nicolas, M.; Wolfer, A.; Raj, K.; Kummer, J.; Mill, P.; van Noort, M.; Hui, C.; Clevers, H.; Dotto, G.; Radtke, F. Notch1 functions as a tumor suppressor in mouse skin. Nat. Genet 2003, 33, 416–421. [Google Scholar]
- Puente, X.S.; Pinyol, M.; Quesada, V.; Conde, L.; Ordóñez, G.R.; Villamor, N.; Escaramis, G.; Jares, P.; Beà, S.; González-Díaz, M.; et al. Whole-genome sequencing identifies recurrent mutations in chronic lymphocytic leukaemia. Nature 2011, 475, 101–105. [Google Scholar] [Green Version]
- Klinakis, A.; Lobry, C.; Abdel-Wahab, O.; Oh, P.; Haeno, H.; Buonamici, S.; van De Walle, I.; Cathelin, S.; Trimarchi, T.; Araldi, E.; et al. A novel tumour-suppressor function for the notch pathway in myeloid leukaemia. Nature 2011, 473, 230–233. [Google Scholar]
- Vodovar, N.; Schweisguth, F. Functions of o-fucosyltransferase in Notch trafficking and signaling: Towards the end of a controversy? J. Biol 2008, 7, 7. [Google Scholar]
- Bray, S.J. Notch signalling: A simple pathway becomes complex. Nat. Rev. Mol. Cell Biol 2006, 7, 678–689. [Google Scholar]
- Logeat, F.; Bessia, C.; Brou, C.; Lebail, O.; Jarriault, S.; Seidah, N.G.; Israël, A. The Notch1 receptor is cleaved constitutively by a furin-like convertase. Proc. Natl. Acad. Sci. USA 1998, 95, 8108–8112. [Google Scholar]
- D’Souza, B.; Meloty-Kapella, L.; Weinmaster, G. Canonical and non-canonical Notch ligands. Curr. Top. Dev. Biol 2010, 92, 73–129. [Google Scholar]
- Gale, N.W.; Dominguez, M.G.; Noguera, I.; Pan, L.; Hughes, V.; Valenzuela, D.M.; Murphy, A.J.; Adams, N.C.; Lin, H.C.; Holash, J.; et al. Haploinsufficiency of Delta-like 4 ligand results in embryonic lethality due to major defects in arterial and vascular development. Proc. Natl. Acad. Sci. USA 2004, 101, 15949–15954. [Google Scholar]
- Xue, Y.; Gao, X.; Lindsell, C.; Norton, C.; Chang, B.; Hicks, C.; Gendron-Maguire, M.; Rand, E.; Weinmaster, G.; Gridley, T. Embryonic lethality and vascular defects in mice lacking the Notch ligand Jagged1. Hum. Mol. Genet 1999, 8, 723–730. [Google Scholar]
- Hrabe de Angelis, M.; McIntyre, J.; Gossler, A. Maintenance of somite borders in mice requires the Delta homologue Dll1. Nature 1997, 386, 717–721. [Google Scholar]
- Jiang, R.; Lan, Y.; Chapman, H.; Shawber, C.; Norton, C.; Serreze, D.; Weinmaster, G.; Gridley, T. Defects in limb, craniofacial, and thymic development in Jagged2 mutant mice. Genes Dev 1998, 12, 1046–1057. [Google Scholar]
- Hozumi, K.; Mailhos, C.; Negishi, N.; Hirano, K.-I.; Yahata, T.; Ando, K.; Zuklys, S.; Holländer, G.A.; Shima, D.T.; Habu, S. Delta-like 4 is indispensable in thymic environment specific for T cell development. J. Exp. Med 2008, 205, 2507–2513. [Google Scholar]
- Koch, U.; Fiorini, E.; Benedito, R.; Besseyrias, V.; Schuster-Gossler, K.; Pierres, M.; Manley, N.R.; Duarte, A.; Macdonald, H.R.; Radtke, F. Delta-like 4 is the essential, nonredundant ligand for Notch1 during thymic T cell lineage commitment. J. Exp. Med 2008, 205, 2515–2523. [Google Scholar]
- Ladi, E.; Nichols, J.; Ge, W.; Miyamoto, A.; Yao, C.; Yang, L.-T.; Boulter, J.; Sun, Y.; Kintner, C.; Weinmaster, G. The divergent DSL ligand Dll3 does not activate Notch signaling but cell autonomously attenuates signaling induced by other DSL ligands. J. Cell Biol 2005, 170, 983–992. [Google Scholar]
- Wu, E.; Croucher, P.; McKie, N. Expression of members of the novel membrane linked metalloproteinase family adam in cells derived from a range of haematological malignancies. Biochem. Biophys. Res. Commun 1997, 235, 437–442. [Google Scholar]
- Van Tetering, G.; van Diest, P.; Verlaan, I.; van der Wall, E.; Kopan, R.; Vooijs, M. Metalloprotease ADAM10 is required for Notch1 site 2 cleavage. J. Biol. Chem 2009, 284, 31018–31027. [Google Scholar]
- Weber, S.; Niessen, M.T.; Prox, J.; Lüllmann-Rauch, R.; Schmitz, A.; Schwanbeck, R.; Blobel, C.P.; Jorissen, E.; de Strooper, B.; Niessen, C.M.; et al. The disintegrin/metalloproteinase ADAM10 is essential for epidermal integrity and Notch-mediated signaling. Development 2011, 138, 495–505. [Google Scholar]
- Brou, C.; Logeat, F.; Gupta, N.; Bessia, C.; LeBail, O.; Doedens, J.; Cumano, A.; Roux, P.; Black, R.; Israël, A. A novel proteolytic cleavage involved in Notch signaling: The role of the disintegrin-metalloprotease TACE. Mol. Cell 2000, 5, 207–216. [Google Scholar]
- Murthy, A.; Shao, Y.W.; Narala, S.R.; Molyneux, S.D.; Zúñiga-Pflücker, J.C.; Khokha, R. Notch activation by the metalloproteinase ADAM17 regulates myeloproliferation and atopic barrier immunity by suppressing epithelial cytokine synthesis. Immunity 2012, 36, 105–119. [Google Scholar]
- Wolfe, M.S. Gamma-secretase in biology and medicine. Semin. Cell Dev. Biol 2009, 20, 219–224. [Google Scholar]
- Hirsinger, E.; Malapert, P.; Dubrulle, J.; Delfini, M.C.; Duprez, D.; Henrique, D.; Ish-Horowicz, D.; Pourquié, O. Notch signalling acts in postmitotic avian myogenic cells to control myoD activation. Development 2001, 128, 107–116. [Google Scholar]
- Kopan, R.; Nye, J.; Weintraub, H. The intracellular domain of mouse Notch: A constitutively activated repressor of myogenesis directed at the basic helix-loop-helix region of myoD. Development 1994, 120, 2385–2396. [Google Scholar]
- Robey, E.; Chang, D.; Itano, A.; Cado, D.; Alexander, H.; Lans, D.; Weinmaster, G.; Salmon, P. An activated form of Notch influences the choice between CD4 and CD8 T cell lineages. Cell 1996, 87, 483–492. [Google Scholar]
- Gaiano, N.; Fishell, G. The role of Notch in promoting glial and neural stem cell fates. Annu. Rev. Neurosci 2002, 25, 471–490. [Google Scholar]
- Bray, S.; Bernard, F. Notch targets and their regulation. Curr. Top. Dev. Biol 2010, 92, 253–275. [Google Scholar]
- High, F.A.; Epstein, J.A. The multifaceted role of Notch in cardiac development and disease. Nat. Rev. Genet 2008, 9, 49–61. [Google Scholar]
- Shimizu, Y.; Okuda-Shimizu, Y.; Hendershot, L.M. Ubiquitylation of an ERAD substrate occurs on multiple types of amino acids. Mol. Cell 2010, 40, 917–926. [Google Scholar]
- Ciechanover, A. Intracellular protein degradation: From a vague idea thru the lysosome and the ubiquitin-proteasome system and onto human diseases and drug targeting. Cell Death Differ 2005, 12, 1178–1190. [Google Scholar]
- Metzger, M.B.; Hristova, V.A.; Weissman, A.M. HECT and RING finger families of E3 ubiquitin ligases at a glance. J. Cell Sci 2012, 125, 531–537. [Google Scholar]
- Amerik, A.; Hochstrasser, M. Mechanism and function of deubiquitinating enzymes. Biochim. Biophys. Acta 2004, 1695, 189. [Google Scholar]
- Komander, D.; Clague, M.J.; Urbé, S. Breaking the chains: Structure and function of the deubiquitinases. Nat. Rev. Mol. Cell Biol 2009, 10, 550–563. [Google Scholar]
- Zhang, J.; Liu, M.; Su, Y.; Du, J.; Zhu, A.J. A targeted in vivo RNAi screen reveals deubiquitinases as new regulators of Notch signaling. G3 2012, 2, 1563–1575. [Google Scholar]
- Lin, Z.; Yang, H.; Kong, Q.; Li, J.; Lee, S.-M.; Gao, B.; Dong, H.; Wei, J.; Song, J.; Zhang, D.D.; et al. USP22 antagonizes p53 transcriptional activation by deubiquitinating Sirt1 to suppress cell apoptosis and is required for mouse embryonic development. Mol. Cell 2012, 46, 484–494. [Google Scholar]
- Chauhan, D.; Tian, Z.; Nicholson, B.; Kumar, K.G.S.; Zhou, B.; Carrasco, R.; Mcdermott, J.L.; Leach, C.A.; Fulcinniti, M.; Kodrasov, M.P.; et al. A small molecule inhibitor of ubiquitin-specific protease-7 induces apoptosis in multiple myeloma cells and overcomes bortezomib resistance. Cancer Cell 2012, 22, 345–358. [Google Scholar]
- Eichhorn, P.J.A.; Rodón, L.; Gonzàlez-Juncà, A.; Dirac, A.; Gili, M.; Martínez-Sáez, E.; Aura, C.; Barba, I.; Peg, V.; Prat, A.; et al. USP15 stabilizes TGF-β receptor I and promotes oncogenesis through the activation of TGF-β signaling in glioblastoma. Nat. Med 2012, 18, 429–435. [Google Scholar]
- Lee, B.-H.; Lee, M.J.; Park, S.; Oh, D.-C.; Elsasser, S.; Chen, P.-C.; Gartner, C.; Dimova, N.; Hanna, J.; Gygi, S.P.; et al. Enhancement of proteasome activity by a small-molecule inhibitor of USP14. Nature 2010, 467, 179–184. [Google Scholar]
- Gupta-Rossi, N.; Le Bail, O.; Gonen, H.; Brou, C.; Logeat, F.; Six, E.; Ciechanover, A.; Israel, A. Functional interaction between Sel-10, an F-box protein, and the nuclear form of activated Notch1 receptor. J. Biol. Chem 2001, 276, 34371–34378. [Google Scholar]
- Oberg, C.; Li, J.; Pauley, A.; Wolf, E.; Gurney, M.; Lendahl, U. The Notch intracellular domain is ubiquitinated and negatively regulated by the mammalian Sel-10 homolog. J. Biol. Chem 2001, 276, 35847–35853. [Google Scholar]
- Wu, G.; Lyapina, S.; Das, I.; Li, J.; Gurney, M.; Pauley, A.; Chui, I.; Deshaies, R.; Kitajewski, J. Sel-10 is an inhibitor of Notch signaling that targets Notch for ubiquitin-mediated protein degradation. Mol. Cell Biol 2001, 21, 7403–7415. [Google Scholar]
- Hubbard, E.; Wu, G.; Kitajewski, J.; Greenwald, I. Sel-10 a negative regulator of lin-12 activity in caenorhabditis elegans, encodes a member of the cdc4 family of proteins. Genes Dev 1997, 11, 3182–3193. [Google Scholar]
- Moberg, K.; Bell, D.; Wahrer, D.; Haber, D.; Hariharan, I. Archipelago regulates Cyclin E levels in Drosophila and is mutated in human cancer cell lines. Nature 2001, 413, 311. [Google Scholar]
- Hao, B.; Oehlmann, S.; Sowa, M.E.; Harper, J.W.; Pavletich, N.P. Structure of a Fbw7-Skp1-Cyclin E complex: Multisite-phosphorylated substrate recognition by SCF ubiquitin ligases. Mol. Cell 2007, 26, 131–143. [Google Scholar]
- Orlicky, S.; Tang, X.; Willems, A.; Tyers, M.; Sicheri, F. Structural basis for phosphodependent substrate selection and orientation by the SCF Cdc4 ubiquitin ligase. Cell 2003, 112, 243–256. [Google Scholar]
- Welcker, M.; Clurman, B.E. Fbw7 ubiquitin ligase: A tumour suppressor at the crossroads of cell division, growth and differentiation. Nat. Rev. Cancer 2008, 8, 83–93. [Google Scholar]
- Fryer, C.J.; White, J.B.; Jones, K.A. Mastermind recruits CycC:Cdk8 to phosphorylate the Notch ICD and coordinate activation with turnover. Mol. Cell 2004, 16, 509–520. [Google Scholar]
- Mao, J.-H.; Perez-Losada, J.; Wu, D.; Delrosario, R.; Tsunematsu, R.; Nakayama, K.I.; Brown, K.; Bryson, S.; Balmain, A. Fbxw7/Cdc4 is a p53-dependent, haploinsufficient tumour suppressor gene. Nature 2004, 432, 775–779. [Google Scholar]
- Yatim, A.; Benne, C.; Sobhian, B.; Laurent-Chabalier, S.; Deas, O.; Judde, J.-G.; Lelievre, J.-D.; Levy, Y.; Benkirane, M. Notch1 nuclear interactome reveals key regulators of its transcriptional activity and oncogenic function. Mol. Cell 2012, 48, 445–458. [Google Scholar]
- Brou, C. Intracellular trafficking of Notch receptors and ligands. Exp. Cell Res 2009, 315, 1549–1555. [Google Scholar]
- Seugnet, L.; Simpson, P.; Haenlin, M. Requirement for Dynamin during Notch signaling in Drosophila neurogenesis. Dev. Biol 1997, 192, 585–598. [Google Scholar]
- McGill, M.A.; Dho, S.E.; Weinmaster, G.; McGlade, C.J. Numb regulates post-endocytic trafficking and degradation of Notch1. J. Biol. Chem 2009, 284, 26427–26438. [Google Scholar]
- Chastagner, P.; Israël, A.; Brou, C. AIP4/Itch regulates Notch receptor degradation in the absence of ligand. PLoS One 2008, 3, e2735. [Google Scholar]
- Sakata, T.; Sakaguchi, H.; Tsuda, L.; Higashitani, A.; Aigaki, T.; Matsuno, K.; Hayashi, S. Drosophila Nedd4 regulates endocytosis of Notch and suppresses its ligand-independent activation. Curr. Biol 2004, 14, 2228–2236. [Google Scholar]
- Vaccari, T.; Lu, H.; Kanwar, R.; Fortini, M.; Bilder, D. Endosomal entry regulates Notch receptor activation in Drosophila melanogaster. J. Cell Biol 2008, 180, 755–762. [Google Scholar]
- Wilkin, M.; Carbery, A.-M.; Fostier, M.; Aslam, H.; Mazaleyrat, S.; Higgs, J.; Myat, A.; Evans, D.; Cornell, M.; Baron, M. Regulation of Notch endosomal sorting and signaling by Drosophila Nedd4 family proteins. Curr. Biol 2004, 14, 2237–2244. [Google Scholar]
- Kaether, C.; Schmitt, S.; Willem, M.; Haass, C. Amyloid precursor protein and Notch intracellular domains are generated after transport of their precursors to the cell surface. Traffic 2006, 7, 408–415. [Google Scholar]
- Henne, W.M.; Buchkovich, N.J.; Emr, S.D. The ESCRT pathway. Dev. Cell 2011, 21, 77–91. [Google Scholar]
- Macdonald, C.; Buchkovich, N.J.; Stringer, D.K.; Emr, S.D.; Piper, R.C. Cargo ubiquitination is essential for multivesicular body intralumenal vesicle formation. EMBO Rep 2012, 13, 331–338. [Google Scholar]
- Moberg, K.; Schelble, S.; Burdick, S.; Hariharan, I. Mutations in Erupted, the Drosophila ortholog of mammalian Tumor susceptibility gene 101, elicit non-cell-autonomous overgrowth. Dev. Cell 2005, 9, 699–710. [Google Scholar]
- Thompson, B.; Mathieu, J.; Sung, H.-H.; Loeser, E.; Rorth, P.; Cohen, S. Tumor suppressor properties of the ESCRT-II complex component Vps25 in drosophila. Dev. Cell 2005, 9, 711–720. [Google Scholar]
- Vaccari, T.; Bilder, D. The Drosophila tumor suppressor Vps25 prevents nonautonomous overproliferation by regulating Notch trafficking. Dev. Cell 2005, 9, 687–698. [Google Scholar]
- Vaccari, T.; Rusten, T.E.; Menut, L.; Nezis, I.P.; Brech, A.; Stenmark, H.; Bilder, D. Comparative analysis of ESCRT-II, ESCRT-II and ESCRT-III function in Drosophila by efficient isolation of ESCRT mutants. J. Cell Sci 2009, 122, 2413–2423. [Google Scholar]
- Jaekel, R.; Klein, T. The Drosophila Notch inhibitor and tumor suppressor gene lethal (2) giant discs encodes a conserved regulator of endosomal trafficking. Dev. Cell 2006, 11, 655–669. [Google Scholar]
- Vaccari, T.; Duchi, S.; Cortese, K.; Tacchetti, C.; Bilder, D. The vacuolar ATPase is required for physiological as well as pathological activation of the Notch receptor. Development 2010, 137, 1825–1832. [Google Scholar]
- Kozik, P.; Hodson, N.A.; Sahlender, D.A.; Simecek, N.; Soromani, C.; Wu, J.; Collinson, L.M.; Robinson, M.S. A human genome-wide screen for regulators of clathrin-coated vesicle formation reveals an unexpected role for the v-ATPase. Nat. Cell Biol 2012, 15, 50–60. [Google Scholar]
- Cornell, M.; Evans, D.A.; Mann, R.; Fostier, M.; Flasza, M.; Monthatong, M.; Artavanis-Tsakonas, S.; Baron, M. The Drosophila melanogaster Suppressor of deltex gene, a regulator of the Notch receptor signaling pathway, is an E3 class ubiquitin ligase. Genetics 1999, 152, 567–576. [Google Scholar]
- Perry, W.; Hustad, C.; Swing, D.; O’Sullivan, T.; Jenkins, N.; Copeland, N. The itchy locus encodes a novel ubiquitin protein ligase that is disrupted in a18H mice. Nat. Genet 1998, 18, 143–146. [Google Scholar]
- Matesic, L.; Haines, D.; Copeland, N.; Jenkins, N. Itch genetically interacts with Notch1 in a mouse autoimmune disease model. Hum. Mol. Genet 2006, 15, 3485–3497. [Google Scholar]
- Beres, B.J.; George, R.; Lougher, E.J.; Barton, M.; Verrelli, B.C.; McGlade, C.J.; Rawls, J.A.; Wilson-Rawls, J. Numb regulates Notch1, but not Notch3, during myogenesis. Mech. Dev 2011, 128, 247–257. [Google Scholar]
- McGill, M.; McGlade, C. Mammalian Numb proteins promote Notch1 receptor ubiquitination and degradation of the Notch1 intracellular domain. J. Biol. Chem 2003, 278, 23196–23203. [Google Scholar]
- Hori, K.; Sen, A.; Kirchhausen, T.; Artavanis-Tsakonas, S. Synergy between the ESCRT-III complex and Deltex defines a ligand-independent Notch signal. J. Cell Biol 2011, 195, 1005–1015. [Google Scholar]
- Mukherjee, A.; Veraksa, A.; Bauer, A.; Rosse, C.; Camonis, J.; Artavanis-Tsakonas, S. Regulation of Notch signalling by non-visual β-arrestin. Nat. Cell Biol 2005, 7, 1191–1201. [Google Scholar]
- Moretti, J.; Chastagner, P.; Liang, C.-C.; Cohn, M.A.; Israel, A.; Brou, C. The Ubiquitin-specific protease 12 (USP12) is a negative regulator of Notch signaling acting on Notch receptor trafficking toward degradation. J. Biol. Chem 2012, 287, 29429–29441. [Google Scholar]
- Diederich, R.; Matsuno, K.; Hing, H.; Artavanis-Tsakonas, S. Cytosolic interaction between Deltex and Notch ankyrin repeats implicates Deltex in the Notch signaling pathway. Development 1994, 120, 473–481. [Google Scholar]
- Hori, K.; Fostier, M.; Ito, M.; Fuwa, T.; Go, M.; Okano, H.; Baron, M.; Matsuno, K. Drosophila Deltex mediates Suppressor of hairless-independent and late-endosomal activation of Notch signaling. Development 2004, 131, 5527–5537. [Google Scholar]
- Wilkin, M.; Tongngok, P.; Gensch, N.; Clemence, S.; Motoki, M.; Yamada, K.; Hori, K.; Taniguchi-Kanai, M.; Franklin, E.; Matsuno, K.; et al. Drosophila HOPS and AP-3 complex genes are required for a Deltex-regulated activation of Notch in the endosomal trafficking pathway. Dev. Cell 2008, 15, 762–772. [Google Scholar]
- Izon, D.J.; Aster, J.C.; He, Y.; Weng, A.; Karnell, F.G.; Patriub, V.; Xu, L.; Bakkour, S.; Rodriguez, C.; Allman, D.; et al. Deltex1 redirects lymphoid progenitors to the B cell lineage by antagonizing Notch1. Immunity 2002, 16, 231–243. [Google Scholar]
- Kishi, N.; Tang, Z.; Maeda, Y.; Hirai, A.; Mo, R.; Ito, M.; Suzuki, S.; Nakao, K.; Kinoshita, T.; Kadesch, T.; et al. Murine homologs of Deltex define a novel gene family involved in vertebrate Notch signaling and neurogenesis. Int. J. Dev. Neurosci 2001, 19, 21–35. [Google Scholar]
- Lehar, S.; Bevan, M. T cells develop normally in the absence of both Deltex1 and Deltex2. Mol. Cell. Biol 2006, 26, 7358–7371. [Google Scholar]
- Allgood, A.G.; Barrick, D. Mapping the Deltex-binding surface on the Notch ankyrin domain using analytical ultracentrifugation. J. Mol. Biol 2011, 414, 243–259. [Google Scholar]
- Zweifel, M.; Leahy, D.; Barrick, D. Structure and Notch receptor binding of the tandem WWEdomain of Deltex. Structure 2005, 13, 1599–1611. [Google Scholar]
- Jackson, P.; Eldridge, A.; Freed, E.; Furstenthal, L.; Hsu, J.; Kaiser, B.; Reimann, J. The lore of the rings: Substrate recognition and catalysis by ubiquitin ligases. Trends Cell Biol 2000, 10, 429–439. [Google Scholar]
- Takeyama, K.; Aguiar, R.; Gu, L.; He, C.; Freeman, G.; Kutok, J.; Aster, J.; Shipp, M. The BAL-binding protein BBAP and related Deltex family members exhibit ubiquitin-protein isopeptide ligase activity. J. Biol. Chem 2003, 278, 21930–21937. [Google Scholar]
- Chastagner, P.; Israel, A.; Brou, C. Itch/AIP4 mediates Deltex degradation through the formation of K29-linked polyubiquitin chains. EMBO Rep 2006, 7, 1147–1153. [Google Scholar]
- Moretti, J.; Chastagner, P.; Gastaldello, S.; Heuss, S.F.; Dirac, A.M.; Bernards, R.; Masucci, M.G.; Israel, A.; Brou, C. The translation initiation factor 3f (eIF3f) exhibits a deubiquitinase activity regulating Notch activation. PLoS Biol 2010, 8, e1000545. [Google Scholar]
- Jehn, B.; Dittert, I.; Beyer, S.; von Der Mark, K.; Bielke, W. C-cbl binding and ubiquitin dependent lysosomal degradation of membrane-associated Notch1. J. Biol. Chem 2002, 277, 8033–8040. [Google Scholar]
- Wang, Y.; Chen, Z.; Bergmann, A. Regulation of EGFR and Notch signaling by distinct isoforms of d-Cbl during Drosophila development. Dev. Biol 2010, 342, 1–10. [Google Scholar]
- Gupta-Rossi, N.; Six, E.; LeBail, O.; Logeat, F.; Chastagner, P.; Olry, A.; Israel, A.; Brou, C. Monoubiquitination and endocytosis direct γ-secretase cleavage of activated Notch receptor. J. Cell Biol 2004, 166, 73–83. [Google Scholar]
- Sczaniecka, M.; Gladstone, K.; Pettersson, S.; McLaren, L.; Huart, A.-S.; Wallace, M. Mdm2 protein-mediated ubiquitination of Numb protein: Identification of a second physiological substrate of Mdm2 that employs a dual-site docking mechanism. J. Biol. Chem 2012, 287, 14052–14068. [Google Scholar]
- Pettersson, S.; Sczaniecka, M.; McLaren, L.; Russell, F.; Gladstone, K.; Hupp, T.; Wallace, M. Non-degradative ubiquitination of the Notch1 receptor by the E3 ligase Mdm2 activates the Notch signalling pathway. Biochem. J 2013, 450, 523–536. [Google Scholar]
- Sun, Y.; Klauzinska, M.; Lake, R.; Lee, J.M.; Santopietro, S.; Raafat, A.; Salomon, D.; Callahan, R.; Artavanis-Tsakonas, S. Trp53 regulates Notch 4 signaling through Mdm2. J. Cell Sci 2011, 124, 1067–1076. [Google Scholar]
- Deblandre, G.; Lai, E.; Kintner, C. Xenopus Neuralized is a ubiquitin ligase that interacts with xDelta1 and regulates Notch signaling. Dev. Cell 2001, 1, 795–806. [Google Scholar]
- Lai, E.; Deblandre, G.; Kintner, C.; Rubin, G. Drosophila Neuralized is a ubiquitin ligase that promotes the internalization and degradation of Delta. Dev. Cell 2001, 1, 783–794. [Google Scholar]
- Lai, E.; Roegiers, F.; Qin, X.; Jan, Y.; Rubin, G. The ubiquitin ligase Drosophila Mind bomb promotes Notch signaling by regulating the localization and activity of Serrate and Delta. Development 2005, 132, 2319–2332. [Google Scholar]
- Le Borgne, R.; Remaud, S.; Hamel, S.; Schweisguth, F. Two distinct E3 ubiquitin ligases have complementary functions in the regulation of Delta and Serrate signaling in Drosophila. PLoS Biol 2005, 3, e96. [Google Scholar]
- Pavlopoulos, E.; Pitsouli, C.; Klueg, K.; Muskavitch, M.; Moschonas, N.; Delidakis, C. Neuralized encodes a peripheral membrane protein involved in Delta signaling and endocytosis. Dev. Cell 2001, 1, 807–816. [Google Scholar]
- Itoh, M.; Kim, C.-H.; Palardy, G.; Oda, T.; Jiang, Y.-J.; Maust, D.; Yeo, S.-Y.; Lorick, K.; Wright, G.J.; Ariza-McNaughton, L.; et al. Mind bomb is a ubiquitin ligase that is essential for efficient activation of notch signaling by Delta. Dev. Cell 2003, 4, 67–82. [Google Scholar]
- Kang, K.; Lee, D.; Hong, S.; Park, S.-G.; Song, M.-R. The E3 ligase Mind bomb-1 (mib1) modulates Delta-Notch signaling to control neurogenesis and gliogenesis in the developing spinal cord. J. Biol. Chem 2013, 288, 2580–2592. [Google Scholar]
- Daskalaki, A.; Shalaby, N.A.; Kux, K.; Tsoumpekos, G.; Tsibidis, G.D.; Muskavitch, M.A.T.; Delidakis, C. Distinct intracellular motifs of Delta mediate its ubiquitylation and activation by Mindbomb1 and Neuralized. J. Cell Biol 2011, 195, 1017–1031. [Google Scholar]
- Benhra, N.; Vignaux, F.; Dussert, A.; Schweisguth, F.; le Borgne, R. Neuralized promotes basal to apical transcytosis of Delta in epithelial cells. Mol. Biol. Cell 2010, 21, 2078–2086. [Google Scholar]
- Song, R.; Koo, B.; Yoon, K.; Yoon, M.; Yoo, K.; Kim, H.; Oh, H.; Kim, Y.; Han, J.; Kim, C.; et al. Neuralized-2 regulates a Notch ligand in cooperation with Mind bomb-1. J. Biol. Chem 2006, 281, 36391–36400. [Google Scholar]
- Koutelou, E.; Sato, S.; Tomomori-Sato, C.; Florens, L.; Swanson, S.; Washburn, M.; Kokkinaki, M.; Conaway, R.; Conaway, J.; Moschonas, N. Neuralized-like 1 (neurl1) targeted to the plasma membrane by N-myristoylation regulates the Notch ligand Jagged1. J. Biol. Chem 2008, 283, 3846–3853. [Google Scholar]
- Koo, B.-K.; Yoon, M.-J.; Yoon, K.-J.; Im, S.-K.; Kim, Y.-Y.; Kim, C.-H.; Suh, P.-G.; Jan, Y.-N.; Kong, Y.-Y. An obligatory role of Mind bomb-1 in Notch signaling of mammalian development. PLoS One 2007, 2, e1221. [Google Scholar]
- Koo, B.-K.; Lim, H.-S.; Song, R.; Yoon, M.-J.; Yoon, K.-J.; Moon, J.-S.; Kim, Y.-W.; Kwon, M.-C.; Yoo, K.-W.; Kong, M.-P.; et al. Mind bomb 1 is essential for generating functional Notch ligands to activate Notch. Development 2005, 132, 3459–3470. [Google Scholar]
- Heuss, S.F.; Ndiaye-Lobry, D.; Six, E.M.; Israël, A.; Logeat, F. The intracellular region of Notch ligands Dll1 and Dll3 regulates their trafficking and signaling activity. Proc. Natl. Acad. Sci. USA 2008, 105, 11212–11217. [Google Scholar] [Green Version]
- Meloty-Kapella, L.; Shergill, B.; Kuon, J.; Botvinick, E.; Weinmaster, G. Notch ligand endocytosis generates mechanical pulling force dependent on dynamin, epsins, and actin. Dev. Cell 2012, 22, 1299–1312. [Google Scholar]
- Nichols, J.; Miyamoto, A.; Olsen, S.; D’Souza, B.; Yao, C.; Weinmaster, G. DSL ligand endocytosis physically dissociates Notch1 heterodimers before activating proteolysis can occur. J. Cell Biol 2007, 176, 445–458. [Google Scholar]
- Parks, A.; Klueg, K.; Stout, J.; Muskavitch, M. Ligand endocytosis drives receptor dissociation and activation in the Notch pathway. Development 2000, 127, 1373–1385. [Google Scholar]
- Shergill, B.; Meloty-Kapella, L.; Musse, A.A.; Weinmaster, G.; Botvinick, E. Optical tweezers studies on Notch: Single-molecule interaction strength is independent of ligand endocytosis. Dev. Cell 2012, 22, 1313–1320. [Google Scholar]
- Overstreet, E.; Fitch, E.; Fischer, J. Fat facets and Liquid facets promote Delta endocytosis and Delta signaling in the signaling cells. Development 2004, 131, 5355–5366. [Google Scholar]
- Wang, W.; Struhl, G. Drosophila Epsin mediates a select endocytic pathway that DSL ligands must enter to activate Notch. Development 2004, 131, 5367–5380. [Google Scholar]
- Xie, X.; Cho, B.; Fischer, J.A. Drosophila Epsin’s role in Notch ligand cells requires three Epsin protein functions: The lipid binding function of the ENTH domain, a single ubiquitin interaction motif, and a subset of the C-terminal protein binding modules. Dev. Biol 2012, 363, 399–412. [Google Scholar]
- Giagtzoglou, N.; Yamamoto, S.; Zitserman, D.; Graves, H.K.; Schulze, K.L.; Wang, H.; Klein, H.; Roegiers, F.; Bellen, H.J. DEHBP1 controls exocytosis and recycling of Delta during asymmetric divisions. J. Cell Biol 2012, 196, 65–83. [Google Scholar]
- Shah, D.K.; Mohtashami, M.; Zuniga-Pflucker, J.C. Role of recycling, Mindbomb1 association, and exclusion from lipid rafts of Delta-like 4 for effective Notch signaling to drive T cell development. J. Immunol 2012, 189, 5797–5808. [Google Scholar]
- Geffers, I.; Serth, K.; Chapman, G.; Jaekel, R.; Schuster-Gossler, K.; Cordes, R.; Sparrow, D.; Kremmer, E.; Dunwoodie, S.; Klein, T.; et al. Divergent functions and distinct localization of the Notch ligands Dll1 and Dll3 in vivo. J. Cell Biol 2007, 178, 465–476. [Google Scholar]
- Zhang, L.; Widau, R.C.; Herring, B.P.; Gallagher, P.J. Delta-like 1-lysine613 regulates Notch signaling. Biochim. Biophys. Acta 2011, 1813, 2036–2043. [Google Scholar]
- Hamel, S.; Fantini, J.; Schweisguth, F. Notch ligand activity is modulated by glycosphingolipid membrane composition in Drosophila melanogaster. J. Cell Biol 2010, 188, 581–594. [Google Scholar]
- Del Álamo, D.; Rouault, H.; Schweisguth, F. Mechanism and significance of cis-inhibition in Notch signalling. Curr. Biol 2011, 21, R40–R47. [Google Scholar]
- Bras, S.L.; Rondanino, C.; Kriegel-Taki, G.; Dussert, A.; Borgne, R.L. Genetic identification of intracellular trafficking regulators involved in Notch-dependent binary cell fate acquisition following asymmetric cell division. J. Cell Sci 2012, 125, 4886–4901. [Google Scholar]
- Saj, A.; Arziman, Z.; Stempfle, D.; van Belle, W.; Sauder, U.; Horn, T.; Dürrenberger, M.; Paro, R.; Boutros, M.; Merdes, G. A combined ex vivo and in vivo RNAi screen for Notch regulators in Drosophila reveals an extensive Notch interaction network. Dev. Cell 2010, 18, 862–876. [Google Scholar]




| Component function | Drosophila | Caenorhabditis elegans | Mammals |
|---|---|---|---|
| Receptor | Notch | LIN-12, GLP-1 | Notch 1–4 |
| Ligand (DSL) | Delta | APX-1, LAG-2, ARG-2, DSL1-7 | Dll 1, 3, 4 |
| Serrate | Jagged 1, 2 | ||
| E3 ubiquitin ligase | Mindbomb1–2 | Mindbomb 1, 2 (skeletrophin) | |
| Neuralized | Neuralized 1, 2 | ||
| Deltex | Deltex 1–4 | ||
| Nedd4, Su(Dx) | WWP-1 | Nedd4, Itch/AIP4 | |
| Archipelago | SEL-10 | Fbw-7/SEL-10 | |
| d-cbl | Cbl | ||
| Mdm2 | |||
| DUB | USP12 (CG 7023) | USP12 | |
| eIF3-S5 (CG 9769) | eIF3f | ||
© 2013 by the authors; licensee MDPI, Basel, Switzerland. This article is an open-access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Moretti, J.; Brou, C. Ubiquitinations in the Notch Signaling Pathway. Int. J. Mol. Sci. 2013, 14, 6359-6381. https://doi.org/10.3390/ijms14036359
Moretti J, Brou C. Ubiquitinations in the Notch Signaling Pathway. International Journal of Molecular Sciences. 2013; 14(3):6359-6381. https://doi.org/10.3390/ijms14036359
Chicago/Turabian StyleMoretti, Julien, and Christel Brou. 2013. "Ubiquitinations in the Notch Signaling Pathway" International Journal of Molecular Sciences 14, no. 3: 6359-6381. https://doi.org/10.3390/ijms14036359
APA StyleMoretti, J., & Brou, C. (2013). Ubiquitinations in the Notch Signaling Pathway. International Journal of Molecular Sciences, 14(3), 6359-6381. https://doi.org/10.3390/ijms14036359