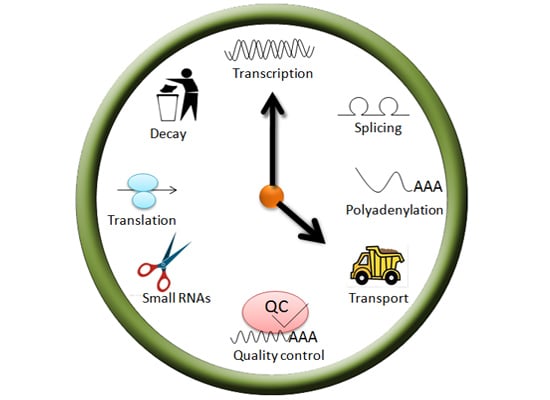
Ribonucleoprotein Complexes That Control Circadian Clocks
Abstract
:1. Introduction
2. Regulation of mRNA Splicing
3. Control of mRNA Polyadenylation
4. mRNA Quality Control
5. mRNA Nuclear Transport
6. Translational Regulation
7. Regulation of mRNA Turnover
8. Summary and Outlook
Acknowledgments
Conflict of Interest
References
- Duguay, D.; Cermakian, N. The crosstalk between physiology and circadian clock proteins. Chronobiol. Int 2009, 26, 1479–1513. [Google Scholar]
- Dunlap, J.C.; Loros, J.J. The neurospora circadian system. J. Biol. Rhythms 2004, 19, 414–424. [Google Scholar]
- Nagel, D.H.; Kay, S.A. Complexity in the wiring and regulation of plant circadian networks. Curr. Biol 2012, 22, R648–R657. [Google Scholar]
- Ozkaya, O.; Rosato, E. The circadian clock of the fly: A neurogenetics journey through time. Adv. Genet 2012, 77, 79–123. [Google Scholar]
- Albrecht, U.; Eichele, G. The mammalian circadian clock. Curr. Opin. Genet. Dev 2003, 13, 271–277. [Google Scholar]
- Bell-Pedersen, D.; Cassone, V.M.; Earnest, D.J.; Golden, S.S.; Hardin, P.E.; Thomas, T.L.; Zoran, M.J. Circadian rhythms from multiple oscillators: Lessons from diverse organisms. Nat. Rev. Genet 2005, 6, 544–556. [Google Scholar]
- Shoguchi, E.; Tanaka, M.; Shinzato, C.; Kawashima, T.; Satoh, N. A genome-wide survey of photoreceptor and circadian genes in the coral, Acropora digitifera. Gene 2013, 515, 426–431. [Google Scholar]
- Chabot, J.R.; Pedraza, J.M.; Luitel, P.; van Oudenaarden, A. Stochastic gene expression out-of-steady-state in the cyanobacterial circadian clock. Nature 2007, 450, 1249–1252. [Google Scholar]
- Feng, D.; Lazar, M.A. Clocks, metabolism, and the epigenome. Mol. Cell 2012, 47, 158–167. [Google Scholar]
- Kawazoe, R.; Mahan, K.M.; Venghaus, B.E.; Carter, M.L.; Herrin, D.L. Circadian regulation of chloroplast transcription in Chlamydomonas is accompanied by little or no fluctuation in RPOD levels or core RNAP activity. Mol. Biol. Rep 2012, 39, 10565–10571. [Google Scholar]
- Lowrey, P.L.; Takahashi, J.S. Mammalian circadian biology: Elucidating genome-wide levels of temporal organization. Annu. Rev. Genomics Hum. Genet 2004, 5, 407–441. [Google Scholar]
- Zwiebel, L.J.; Hardin, P.E.; Liu, X.; Hall, J.C.; Rosbash, M. A post-transcriptional mechanism contributes to circadian cycling of a per-beta-galactosidase fusion protein. Proc. Natl. Acad. Sci. USA 1991, 88, 3882–3886. [Google Scholar]
- So, W.V.; Rosbash, M. Post-transcriptional regulation contributes to Drosophila clock gene mRNA cycling. EMBO J 1997, 16, 7146–7155. [Google Scholar]
- Zhang, L.; Weng, W.; Guo, J. Posttranscriptional mechanisms in controlling eukaryotic circadian rhythms. FEBS Lett 2011, 585, 1400–1405. [Google Scholar]
- Koike, N.; Yoo, S.H.; Huang, H.C.; Kumar, V.; Lee, C.; Kim, T.K.; Takahashi, J.S. Transcriptional architecture and chromatin landscape of the core circadian clock in mammals. Science 2012, 338, 349–354. [Google Scholar]
- Menet, J.S.; Rodriguez, J.; Abruzzi, K.C.; Rosbash, M. Nascent-Seq reveals novel features of mouse circadian transcriptional regulation. ELife Sci 2012, 1, e00011. [Google Scholar]
- Dreyfuss, G.; Kim, V.N.; Kataoka, N. Messenger-RNA-binding proteins and the messages they carry. Nat. Rev. Mol. Cell. Biol 2002, 3, 195–205. [Google Scholar]
- Glisovic, T.; Bachorik, J.L.; Yong, J.; Dreyfuss, G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett 2008, 582, 1977–1986. [Google Scholar]
- Gao, F.B. Understanding fragile X syndrome: Insights from retarded flies. Neuron 2002, 34, 859–862. [Google Scholar]
- Harms, E.; Kivimae, S.; Young, M.W.; Saez, L. Posttranscriptional and posttranslational regulation of clock genes. J. Biol. Rhythms 2004, 19, 361–373. [Google Scholar]
- Cibois, M.; Gautier-Courteille, C.; Legagneux, V.; Paillard, L. Post-transcriptional controls—Adding a new layer of regulation to clock gene expression. Trends Cell Biol 2010, 20, 533–541. [Google Scholar]
- Kojima, S.; Shingle, D.L.; Green, C.B. Post-transcriptional control of circadian rhythms. J. Cell Sci 2011, 124, 311–320. [Google Scholar]
- Staiger, D.; Green, R. RNA-based regulation in the plant circadian clock. Trends Plant Sci 2011, 16, 517–523. [Google Scholar]
- Staiger, D.; Koster, T. Spotlight on post-transcriptional control in the circadian system. Cell. Mol. Life Sci 2011, 68, 71–83. [Google Scholar]
- Kojima, S.; Sher-Chen, E.L.; Green, C.B. Circadian control of mRNA polyadenylation dynamics regulates rhythmic protein expression. Genes Dev 2012, 26, 2724–2736. [Google Scholar]
- Wahl, M.C.; Will, C.L.; Luhrmann, R. The spliceosome: Design principles of a dynamic RNP machine. Cell 2009, 136, 701–718. [Google Scholar]
- Majercak, J.; Chen, W.F.; Edery, I. Splicing of the period gene 3′-terminal intron is regulated by light, circadian clock factors, and phospholipase C. Mol. Cell. Biol 2004, 24, 3359–3372. [Google Scholar]
- Low, K.H.; Chen, W.F.; Yildirim, E.; Edery, I. Natural Variation in the Drosophila melanogaster Clock Gene Period Modulates Splicing of Its 3′-Terminal Intron and Mid-Day Siesta. PLoS One 2012, 7, e49536. [Google Scholar]
- Liu, Y.; Garceau, N.Y.; Loros, J.J.; Dunlap, J.C. Thermally regulated translational control of FRQ mediates aspects of temperature responses in the neurospora circadian clock. Cell 1997, 89, 477–486. [Google Scholar]
- Liu, Y.; Merrow, M.; Loros, J.J.; Dunlap, J.C. How temperature changes reset a circadian oscillator. Science 1998, 281, 825–829. [Google Scholar]
- Neiss, A.; Schafmeier, T.; Brunner, M. Transcriptional regulation and function of the Neurospora clock gene white collar 2 and its isoforms. EMBO Rep 2008, 9, 788–794. [Google Scholar]
- Diernfellner, A.; Colot, H.V.; Dintsis, O.; Loros, J.J.; Dunlap, J.C.; Brunner, M. Long and short isoforms of Neurospora clock protein FRQ support temperature-compensated circadian rhythms. FEBS Lett 2007, 581, 5759–5764. [Google Scholar]
- Collett, M.A.; Dunlap, J.C.; Loros, J.J. Circadian clock-specific roles for the light response protein WHITE COLLAR-2. Mol. Cell. Biol 2001, 21, 2619–2628. [Google Scholar]
- Diernfellner, A.C.; Schafmeier, T.; Merrow, M.W.; Brunner, M. Molecular mechanism of temperature sensing by the circadian clock of Neurospora crassa. Genes Dev 2005, 19, 1968–1973. [Google Scholar]
- Kaldi, K.; Gonzalez, B.H.; Brunner, M. Transcriptional regulation of the Neurospora circadian clock gene wc-1 affects the phase of circadian output. EMBO Rep 2006, 7, 199–204. [Google Scholar]
- Baker, C.L.; Loros, J.J.; Dunlap, J.C. The circadian clock of Neurospora crassa. FEMS Microbiol. Rev 2012, 36, 95–110. [Google Scholar]
- Colot, H.V.; Loros, J.J.; Dunlap, J.C. Temperature-modulated alternative splicing and promoter use in the Circadian clock gene frequency. Mol. Biol. Cell 2005, 16, 5563–5571. [Google Scholar]
- Park, M.J.; Seo, P.J.; Park, C.M. CCA1 alternative splicing as a way of linking the circadian clock to temperature response in Arabidopsis. Plant Signal. Behav 2012, 7, 1194–1196. [Google Scholar]
- Wang, X.; Wu, F.; Xie, Q.; Wang, H.; Wang, Y.; Yue, Y.; Gahura, O.; Ma, S.; Liu, L.; Cao, Y.; et al. SKIP is a component of the spliceosome linking alternative splicing and the circadian clock in Arabidopsis. Plant Cell 2012, 24, 3278–3295. [Google Scholar]
- Jones, M.A.; Williams, B.A.; McNicol, J.; Simpson, C.G.; Brown, J.W.; Harmer, S.L. Mutation of Arabidopsis SPLICEOSOMAL TIMEKEEPER LOCUS1 Causes Circadian Clock Defects. Plant Cell 2012, 24, 4066–4082. [Google Scholar] [Green Version]
- Sanchez, S.E.; Petrillo, E.; Beckwith, E.J.; Zhang, X.; Rugnone, M.L.; Hernando, C.E.; Cuevas, J.C.; Godoy Herz, M.A.; Depetris-Chauvin, A.; Simpson, C.G.; et al. A methyl transferase links the circadian clock to the regulation of alternative splicing. Nature 2010, 468, 112–116. [Google Scholar]
- Hong, S.; Song, H.R.; Lutz, K.; Kerstetter, R.A.; Michael, T.P.; McClung, C.R. Type II protein arginine methyltransferase 5 (PRMT5) is required for circadian period determination in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 2010, 107, 21211–21216. [Google Scholar]
- Shav-Tal, Y.; Zipori, D. PSF and p54nrb/NonO—Multi-functional nuclear proteins. FEBS Lett 2002, 531, 109–114. [Google Scholar]
- Guillaumond, F.; Boyer, B.; Becquet, D.; Guillen, S.; Kuhn, L.; Garin, J.; Belghazi, M.; Bosler, O.; Franc, J.L.; Francois-Bellan, A.M. Chromatin remodeling as a mechanism for circadian prolactin transcription: Rhythmic NONO and SFPQ recruitment to HLTF. FASEB J 2011, 25, 2740–2756. [Google Scholar]
- Peng, R.; Dye, B.T.; Perez, I.; Barnard, D.C.; Thompson, A.B.; Patton, J.G. PSF and p54nrb bind a conserved stem in U5 snRNA. RNA 2002, 8, 1334–1347. [Google Scholar]
- Brown, S.A.; Ripperger, J.; Kadener, S.; Fleury-Olela, F.; Vilbois, F.; Rosbash, M.; Schibler, U. PERIOD1-associated proteins modulate the negative limb of the mammalian circadian oscillator. Science 2005, 308, 693–696. [Google Scholar]
- Passon, D.M.; Lee, M.; Rackham, O.; Stanley, W.A.; Sadowska, A.; Filipovska, A.; Fox, A.H.; Bond, C.S. Structure of the heterodimer of human NONO and paraspeckle protein component 1 and analysis of its role in subnuclear body formation. Proc. Natl. Acad. Sci. USA 2012, 109, 4846–4850. [Google Scholar]
- Kowalska, E.; Ripperger, J.A.; Hoegger, D.C.; Bruegger, P.; Buch, T.; Birchler, T.; Mueller, A.; Albrecht, U.; Contaldo, C.; Brown, S.A. NONO couples the circadian clock to the cell cycle. Proc. Natl. Acad. Sci. USA 2012, 110, 1592–1599. [Google Scholar]
- Jacobson, A.; Peltz, S.W. Interrelationships of the pathways of mRNA decay and translation in eukaryotic cells. Annu. Rev. Biochem 1996, 65, 693–739. [Google Scholar]
- Lin, C.L.; Evans, V.; Shen, S.; Xing, Y.; Richter, J.D. The nuclear experience of CPEB: Implications for RNA processing and translational control. RNA 2010, 16, 338–348. [Google Scholar]
- Kowalska, E.; Ripperger, J.A.; Muheim, C.; Maier, B.; Kurihara, Y.; Fox, A.H.; Kramer, A.; Brown, S.A. Distinct Roles of DBHS Family Members in the Circadian Transcriptional Feedback Loop. Mol. Cell. Biol 2012, 32, 4585–4594. [Google Scholar]
- Green, C.B.; Besharse, J.C. Identification of a novel vertebrate circadian clock-regulated gene encoding the protein nocturnin. Proc. Natl. Acad. Sci. USA 1996, 93, 14884–14888. [Google Scholar]
- Baggs, J.E.; Green, C.B. Nocturnin, a deadenylase in Xenopus laevis retina: A mechanism for posttranscriptional control of circadian-related mRNA. Curr. Biol 2003, 13, 189–198. [Google Scholar]
- Wang, Y.; Osterbur, D.L.; Megaw, P.L.; Tosini, G.; Fukuhara, C.; Green, C.B.; Besharse, J.C. Rhythmic expression of Nocturnin mRNA in multiple tissues of the mouse. BMC Dev. Biol. 2001, 1. [Google Scholar] [CrossRef]
- Oishi, K.; Miyazaki, K.; Kadota, K.; Kikuno, R.; Nagase, T.; Atsumi, G.; Ohkura, N.; Azama, T.; Mesaki, M.; Yukimasa, S.; et al. Genome-wide expression analysis of mouse liver reveals CLOCK-regulated circadian output genes. J. Biol. Chem 2003, 278, 41519–41527. [Google Scholar]
- Liu, X.; Green, C.B. A novel promoter element, photoreceptor conserved element II, directs photoreceptor-specific expression of nocturnin in Xenopus laevis. J. Biol. Chem 2001, 276, 15146–15154. [Google Scholar]
- Li, R.; Yue, J.; Zhang, Y.; Zhou, L.; Hao, W.; Yuan, J.; Qiang, B.; Ding, J.M.; Peng, X.; Cao, J.M. CLOCK/BMAL1 regulates human nocturnin transcription through binding to the E-box of nocturnin promoter. Mol. Cell. Biochem 2008, 317, 169–177. [Google Scholar]
- Hussain, M.M.; Pan, X. Clock regulation of dietary lipid absorption. Curr. Opin. Clin. Nutr. Metab. Care 2012, 15, 336–341. [Google Scholar]
- Muller, W.E.; Wang, X.; Grebenjuk, V.A.; Korzhev, M.; Wiens, M.; Schlossmacher, U.; Schroder, H.C. Nocturnin in the demosponge Suberites domuncula: A potential circadian clock protein controlling glycogenin synthesis in sponges. Biochem. J 2012, 448, 233–242. [Google Scholar]
- Aktan, F. iNOS-mediated nitric oxide production and its regulation. Life Sci 2004, 75, 639–653. [Google Scholar]
- Niu, S.; Shingle, D.L.; Garbarino-Pico, E.; Kojima, S.; Gilbert, M.; Green, C.B. The circadian deadenylase Nocturnin is necessary for stabilization of the iNOS mRNA in mice. PLoS One 2011, 6, e26954. [Google Scholar]
- Cheng, P.; He, Q.; He, Q.; Wang, L.; Liu, Y. Regulation of the Neurospora circadian clock by an RNA helicase. Genes Dev 2005, 19, 234–241. [Google Scholar]
- Guo, J.; Cheng, P.; Yuan, H.; Liu, Y. The Exosome regulates circadian gene expression in a posttranscriptional negative feedback loop. Cell 2009, 138, 1236–1246. [Google Scholar]
- Guo, J.; Cheng, P.; Liu, Y. Functional significance of FRH in regulating the phosphorylation and stability of Neurospora circadian clock protein FRQ. J. Biol. Chem 2010, 285, 11508–11515. [Google Scholar]
- Hunt, S.M.; Thompson, S.; Elvin, M.; Heintzen, C. VIVID interacts with the WHITE COLLAR complex and FREQUENCY-interacting RNA helicase to alter light and clock responses in Neurospora. Proc. Natl. Acad. Sci. USA 2010, 107, 16709–16714. [Google Scholar]
- Hunt, S.; Elvin, M.; Heintzen, C. Temperature-sensitive and circadian oscillators of Neurospora crassa share components. Genetics 2012, 191, 119–131. [Google Scholar]
- Shi, M.; Collett, M.; Loros, J.J.; Dunlap, J.C. FRQ-interacting RNA helicase mediates negative and positive feedback in the Neurospora circadian clock. Genetics 2010, 184, 351–361. [Google Scholar]
- Belden, W.J.; Lewis, Z.A.; Selker, E.U.; Loros, J.J.; Dunlap, J.C. CHD1 remodels chromatin and influences transient DNA methylation at the clock gene frequency. PLoS Genet 2011, 7, e1002166. [Google Scholar]
- Van Hoof, A.; Lennertz, P.; Parker, R. Yeast exosome mutants accumulate 3′-extended polyadenylated forms of U4 small nuclear RNA and small nucleolar RNAs. Mol. Cell. Biol 2000, 20, 441–452. [Google Scholar]
- LaCava, J.; Houseley, J.; Saveanu, C.; Petfalski, E.; Thompson, E.; Jacquier, A.; Tollervey, D. RNA degradation by the exosome is promoted by a nuclear polyadenylation complex. Cell 2005, 121, 713–724. [Google Scholar]
- Holub, P.; Lalakova, J.; Cerna, H.; Pasulka, J.; Sarazova, M.; Hrazdilova, K.; Arce, M.S.; Hobor, F.; Stefl, R.; Vanacova, S. Air2p is critical for the assembly and RNA-binding of the TRAMP complex and the KOW domain of Mtr4p is crucial for exosome activation. Nucleic Acids Res 2012, 40, 5679–5693. [Google Scholar]
- Fasken, M.B.; Leung, S.W.; Banerjee, A.; Kodani, M.O.; Chavez, R.; Bowman, E.A.; Purohit, M.K.; Rubinson, M.E.; Rubinson, E.H.; Corbett, A.H. Air1 zinc knuckles 4 and 5 and a conserved IWRXY motif are critical for the function and integrity of the Trf4/5-Air1/2-Mtr4 polyadenylation (TRAMP) RNA quality control complex. J. Biol. Chem 2011, 286, 37429–37445. [Google Scholar]
- Sadoff, B.U.; Heath-Pagliuso, S.; Castano, I.B.; Zhu, Y.; Kieff, F.S.; Christman, M.F. Isolation of mutants of Saccharomyces cerevisiae requiring DNA topoisomerase I. Genetics 1995, 141, 465–479. [Google Scholar]
- Haracska, L.; Johnson, R.E.; Prakash, L.; Prakash, S. Trf4 and Trf5 proteins of Saccharomyces cerevisiae exhibit poly(A) RNA polymerase activity but no DNA polymerase activity. Mol. Cell. Biol 2005, 25, 10183–10189. [Google Scholar]
- Wagner, E.; Lykke-Andersen, J. mRNA surveillance: The perfect persist. J. Cell Sci 2002, 115, 3033–3038. [Google Scholar]
- Doma, M.K.; Parker, R. RNA quality control in eukaryotes. Cell 2007, 131, 660–668. [Google Scholar]
- He, F.; Li, X.; Spatrick, P.; Casillo, R.; Dong, S.; Jacobson, A. Genome-wide analysis of mRNAs regulated by the nonsense-mediated and 5′ to 3′ mRNA decay pathways in yeast. Mol. Cell 2003, 12, 1439–1452. [Google Scholar]
- Rehwinkel, J.; Letunic, I.; Raes, J.; Bork, P.; Izaurralde, E. Nonsense-mediated mRNA decay factors act in concert to regulate common mRNA targets. RNA 2005, 11, 1530–1544. [Google Scholar]
- Chang, Y.F.; Imam, J.S.; Wilkinson, M.F. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem 2007, 76, 51–74. [Google Scholar]
- Metzstein, M.M.; Krasnow, M.A. Functions of the nonsense-mediated mRNA decay pathway in Drosophila development. PLoS Genet 2006, 2, e180. [Google Scholar]
- Rebbapragada, I.; Lykke-Andersen, J. Execution of nonsense-mediated mRNA decay: What defines a substrate? Curr. Opin. Cell. Biol 2009, 21, 394–402. [Google Scholar]
- Morgan, L.W.; Feldman, J.F. Isolation and characterization of a temperature-sensitive circadian clock mutant of Neurospora crassa. Genetics 1997, 146, 525–530. [Google Scholar]
- Jeong, H.J.; Kim, Y.J.; Kim, S.H.; Kim, Y.H.; Lee, I.J.; Kim, Y.K.; Shin, J.S. Nonsense-mediated mRNA decay factors, UPF1 and UPF3, contribute to plant defense. Plant Cell Physiol 2011, 52, 2147–2156. [Google Scholar]
- Shi, C.; Baldwin, I.T.; Wu, J. Arabidopsis plants having defects in nonsense-mediated mRNA decay factors UPF1, UPF2, and UPF3 show photoperiod-dependent phenotypes in development and stress responses. J. Integr. Plant Biol 2012, 54, 99–114. [Google Scholar]
- Heintzen, C.; Nater, M.; Apel, K.; Staiger, D. AtGRP7, a nuclear RNA-binding protein as a component of a circadian-regulated negative feedback loop in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 1997, 94, 8515–8120. [Google Scholar]
- Schoning, J.C.; Streitner, C.; Page, D.R.; Hennig, S.; Uchida, K.; Wolf, E.; Furuya, M.; Staiger, D. Auto-regulation of the circadian slave oscillator component AtGRP7 and regulation of its targets is impaired by a single RNA recognition motif point mutation. Plant J 2007, 52, 1119–1130. [Google Scholar]
- Staiger, D.; Zecca, L.; Wieczorek Kirk, D.A.; Apel, K.; Eckstein, L. The circadian clock regulated RNA-binding protein AtGRP7 autoregulates its expression by influencing alternative splicing of its own pre-mRNA. Plant J 2003, 33, 361–371. [Google Scholar]
- Kim, J.S.; Jung, H.J.; Lee, H.J.; Kim, K.A.; Goh, C.H.; Woo, Y.; Oh, S.H.; Han, Y.S.; Kang, H. Glycine-rich RNA-binding protein 7 affects abiotic stress responses by regulating stomata opening and closing in Arabidopsis thaliana. Plant J 2008, 55, 455–466. [Google Scholar]
- Streitner, C.; Hennig, L.; Korneli, C.; Staiger, D. Global transcript profiling of transgenic plants constitutively overexpressing the RNA-binding protein AtGRP7. BMC Plant Biol. 2010, 10. [Google Scholar] [CrossRef]
- Kelly, S.M.; Corbett, A.H. Messenger RNA export from the nucleus: A series of molecular wardrobe changes. Traffic 2009, 10, 1199–1208. [Google Scholar]
- Morf, J.; Rey, G.; Schneider, K.; Stratmann, M.; Fujita, J.; Naef, F.; Schibler, U. Cold-inducible RNA-binding protein modulates circadian gene expression posttranscriptionally. Science 2012, 338, 379–383. [Google Scholar]
- Xia, Z.P.; Zheng, X.M.; Zheng, H.; Liu, X.J.; Liu, G.Y.; Wang, X.H. Downregulation of cold-inducible RNA-binding protein activates mitogen-activated protein kinases and impairs spermatogenic function in mouse testes. Asian J. Androl 2012, 14, 884–889. [Google Scholar]
- Inoue, S.; Shimoda, M.; Nishinokubi, I.; Siomi, M.C.; Okamura, M.; Nakamura, A.; Kobayashi, S.; Ishida, N.; Siomi, H. A role for the Drosophila fragile X-related gene in circadian output. Curr. Biol 2002, 12, 1331–1335. [Google Scholar]
- Dockendorff, T.C.; Su, H.S.; McBride, S.M.; Yang, Z.; Choi, C.H.; Siwicki, K.K.; Sehgal, A.; Jongens, T.A. Drosophila lacking dfmr1 activity show defects in circadian output and fail to maintain courtship interest. Neuron 2002, 34, 973–984. [Google Scholar]
- Zhang, J.; Fang, Z.; Jud, C.; Vansteensel, M.J.; Kaasik, K.; Lee, C.C.; Albrecht, U.; Tamanini, F.; Meijer, J.H.; Oostra, B.A.; et al. Fragile X-related proteins regulate mammalian circadian behavioral rhythms. Am. J. Hum. Genet 2008, 83, 43–52. [Google Scholar]
- Liang, S.; Hitomi, M.; Hu, Y.H.; Liu, Y.; Tartakoff, A.M. A DEAD-box-family protein is required for nucleocytoplasmic transport of yeast mRNA. Mol.Cell. Biol 1996, 16, 5139–5146. [Google Scholar]
- Reddy, A.B.; Karp, N.A.; Maywood, E.S.; Sage, E.A.; Deery, M.; O’Neill, J.S.; Wong, G.K.; Chesham, J.; Odell, M.; Lilley, K.S.; et al. Circadian orchestration of the hepatic proteome. Curr. Biol 2006, 16, 1107–1115. [Google Scholar]
- Jouffe, C.; Cretenet, G.; Symul, L.; Martin, E.; Atger, F.; Naef, F.; Gachon, F. The circadian clock coordinates ribosome biogenesis. PLoS Biol 2013, 11, e1001455. [Google Scholar]
- Newby, L.M.; Jackson, F.R. Regulation of a specific circadian clock output pathway by lark, a putative RNA-binding protein with repressor activity. J. Neurobiol 1996, 31, 117–128. [Google Scholar]
- Sofola, O.; Sundram, V.; Ng, F.; Kleyner, Y.; Morales, J.; Botas, J.; Jackson, F.R.; Nelson, D.L. The Drosophila FMRP and LARK RNA-binding proteins function together to regulate eye development and circadian behavior. J. Neurosci 2008, 28, 10200–10205. [Google Scholar]
- Kojima, S.; Matsumoto, K.; Hirose, M.; Shimada, M.; Nagano, M.; Shigeyoshi, Y.; Hoshino, S.; Ui-Tei, K.; Saigo, K.; Green, C.B.; et al. LARK activates posttranscriptional expression of an essential mammalian clock protein, PERIOD1. Proc. Natl. Acad. Sci. USA 2007, 104, 1859–1864. [Google Scholar]
- Kim, D.Y.; Woo, K.C.; Lee, K.H.; Kim, T.D.; Kim, K.T. hnRNP Q and PTB modulate the circadian oscillation of mouse Rev-erb alpha via IRES-mediated translation. Nucleic Acids Res 2010, 38, 7068–7078. [Google Scholar]
- Kim, T.D.; Woo, K.C.; Cho, S.; Ha, D.C.; Jang, S.K.; Kim, K.T. Rhythmic control of AANAT translation by hnRNP Q in circadian melatonin production. Genes Dev 2007, 21, 797–810. [Google Scholar]
- Lee, K.H.; Woo, K.C.; Kim, D.Y.; Kim, T.D.; Shin, J.; Park, S.M.; Jang, S.K.; Kim, K.T. Rhythmic Interaction between Period1 mRNA and hnRNP Q Leads to Circadian Time-Dependent Translation. Mol. Cell. Biol 2012, 32, 717–728. [Google Scholar]
- Morse, D.; Milos, P.M.; Roux, E.; Hastings, J.W. Circadian regulation of bioluminescence in Gonyaulax involves translational control. Proc. Natl. Acad. Sci. USA 1989, 86, 172–176. [Google Scholar]
- Mittag, M.; Lee, D.H.; Hastings, J.W. Circadian expression of the luciferin-binding protein correlates with the binding of a protein to the 3′ untranslated region of its mRNA. Proc. Natl. Acad. Sci. USA 1994, 91, 5257–5261. [Google Scholar]
- Mittag, M.; Waltenberger, H. In vitro mutagenesis of binding site elements for the clock-controlled proteins CCTR and Chlamy 1. Biol. Chem 1997, 378, 1167–1170. [Google Scholar]
- Kimchi-Sarfaty, C.; Oh, J.M.; Kim, I.W.; Sauna, Z.E.; Calcagno, A.M.; Ambudkar, S.V.; Gottesman, M.M. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science 2007, 315, 525–528. [Google Scholar]
- Zhou, M.; Guo, J.; Cha, J.; Chase, M.; Chen, S.; Barral, J.M.; Sachs, M.S.; Liu, Y. Non-optimal codon usage determines the expression, structure and function of a circadian clock protein. Nature 2013, 495, 111–115. [Google Scholar]
- Xu, Y.; Ma, P.; Shah, P.; Rokas, A.; Liu, Y.; Johnson, C.H. Non-optimal codon usage is a mechanism to achieve circadian clock conditionality. Nature 2013, 495, 116–120. [Google Scholar]
- Carpen, J.D.; von Schantz, M.; Smits, M.; Skene, D.J.; Archer, S.N. A silent polymorphism in the PER1 gene associates with extreme diurnal preference in humans. J. Hum. Genet 2006, 51, 1122–1125. [Google Scholar] [Green Version]
- Parker, R.; Song, H. The enzymes and control of eukaryotic mRNA turnover. Nat. Struct. Mol. Biol 2004, 11, 121–127. [Google Scholar]
- Dziembowski, A.; Lorentzen, E.; Conti, E.; Seraphin, B. A single subunit, Dis3, is essentially responsible for yeast exosome core activity. Nat. Struct. Mol. Biol 2007, 14, 15–22. [Google Scholar]
- Hartung, S.; Hopfner, K.P. Lessons from structural and biochemical studies on the archaeal exosome. Biochem. Soc. Trans 2009, 37, 83–87. [Google Scholar]
- Chen, C.Y.; Shyu, A.B. AU-rich elements: Characterization and importance in mRNA degradation. Trends Biochem. Sci 1995, 20, 465–470. [Google Scholar]
- Benjamin, D.; Schmidlin, M.; Min, L.; Gross, B.; Moroni, C. BRF1 protein turnover and mRNA decay activity are regulated by protein kinase B at the same phosphorylation sites. Mol. Cell. Biol 2006, 26, 9497–9507. [Google Scholar]
- Kim, T.D.; Kim, J.S.; Kim, J.H.; Myung, J.; Chae, H.D.; Woo, K.C.; Jang, S.K.; Koh, D.S.; Kim, K.T. Rhythmic Serotonin N-Acetyltransferase mRNA degradation is essential for the maintenance of its circadian oscillation. Mol. Cell. Biol 2005, 25, 3232–3246. [Google Scholar]
- Kim, D.Y.; Kwak, E.; Kim, S.H.; Lee, K.H.; Woo, K.C.; Kim, K.T. hnRNP Q mediates a phase-dependent translation-coupled mRNA decay of mouse Period3. Nucleic Acids Res 2011, 39, 8901–8914. [Google Scholar]
- Bartel, D.P. MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell 2004, 116, 281–297. [Google Scholar]
- Hammond, S.M. Dicing and slicing: The core machinery of the RNA interference pathway. FEBS Lett 2005, 579, 5822–5829. [Google Scholar]
- Jaskiewicz, L.; Filipowicz, W. Role of Dicer in posttranscriptional RNA silencing. Curr. Top. Microbiol. Immunol 2008, 320, 77–97. [Google Scholar]
- Cheng, H.Y.; Papp, J.W.; Varlamova, O.; Dziema, H.; Russell, B.; Curfman, J.P.; Nakazawa, T.; Shimizu, K.; Okamura, H.; Impey, S.; et al. microRNA modulation of circadian-clock period and entrainment. Neuron 2007, 54, 813–829. [Google Scholar]
- Gatfield, D.; le Martelot, G.; Vejnar, C.E.; Gerlach, D.; Schaad, O.; Fleury-Olela, F.; Ruskeepaa, A.L.; Oresic, M.; Esau, C.C.; Zdobnov, E.M.; et al. Integration of microRNA miR-122 in hepatic circadian gene expression. Genes Dev 2009, 23, 1313–1326. [Google Scholar]
- Kadener, S.; Menet, J.S.; Sugino, K.; Horwich, M.D.; Weissbein, U.; Nawathean, P.; Vagin, V.V.; Zamore, P.D.; Nelson, S.B.; Rosbash, M. A role for microRNAs in the Drosophila circadian clock. Genes Dev 2009, 23, 2179–2191. [Google Scholar]
- Pegoraro, M.; Tauber, E. The role of microRNAs (miRNA) in circadian rhythmicity. J. Genet 2008, 87, 505–511. [Google Scholar]
- Saus, E.; Soria, V.; Escaramis, G.; Vivarelli, F.; Crespo, J.M.; Kagerbauer, B.; Menchon, J.M.; Urretavizcaya, M.; Gratacos, M.; Estivill, X. Genetic variants and abnormal processing of pre-miR-182, a circadian clock modulator, in major depression patients with late insomnia. Hum. Mol. Genet 2010, 19, 4017–4025. [Google Scholar]
- Xu, S.; Witmer, P.D.; Lumayag, S.; Kovacs, B.; Valle, D. MicroRNA (miRNA) transcriptome of mouse retina and identification of a sensory organ-specific miRNA cluster. J. Biol. Chem 2007, 282, 25053–25066. [Google Scholar]
- Zhou, W.; Li, Y.; Wang, X.; Wu, L.; Wang, Y. MiR-206-mediated dynamic mechanism of the mammalian circadian clock. BMC Syst. Biol 2011, 5, 141. [Google Scholar]


© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wang, D.; Liang, X.; Chen, X.; Guo, J. Ribonucleoprotein Complexes That Control Circadian Clocks. Int. J. Mol. Sci. 2013, 14, 9018-9036. https://doi.org/10.3390/ijms14059018
Wang D, Liang X, Chen X, Guo J. Ribonucleoprotein Complexes That Control Circadian Clocks. International Journal of Molecular Sciences. 2013; 14(5):9018-9036. https://doi.org/10.3390/ijms14059018
Chicago/Turabian StyleWang, Dongni, Xiaodi Liang, Xianyun Chen, and Jinhu Guo. 2013. "Ribonucleoprotein Complexes That Control Circadian Clocks" International Journal of Molecular Sciences 14, no. 5: 9018-9036. https://doi.org/10.3390/ijms14059018
APA StyleWang, D., Liang, X., Chen, X., & Guo, J. (2013). Ribonucleoprotein Complexes That Control Circadian Clocks. International Journal of Molecular Sciences, 14(5), 9018-9036. https://doi.org/10.3390/ijms14059018