Induced Production of 1-Methoxy-indol-3-ylmethyl Glucosinolate by Jasmonic Acid and Methyl Jasmonate in Sprouts and Leaves of Pak Choi (Brassica rapa ssp. chinensis)
Abstract
:1. Introduction
2. Results and Discussion
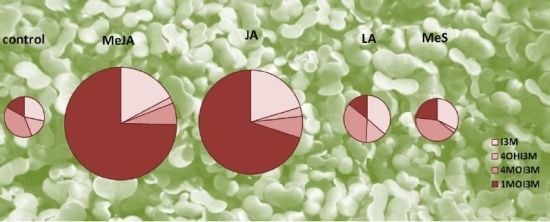
2.1. Indole Glucosinolates Are Induced in Pak Choi Sprouts Treated with Signaling Molecules
2.2. Ontogenetic Differences in Induction of Indole Glucosinolates in Pak Choi
2.3. High Levels of Particular Indole Glucosinolate Breakdown Products after Tissue Disruption of Pak Choi Leaves
3. Experimental Section
3.1. Plant Material
3.2. Elicitors and Plant Treatment
3.3. Sample Preparation and Desulfo-Glucosinolate Analysis by HPLC
3.4. Gene Expression Analysis by Semi-Quantitative Realtime RT-PCR
3.5. Analysis of Breakdown Products of 1-Methoxy-indol-3-ylmethyl Glucosinolate
3.6. Statistical Analysis
4. Conclusions
Supplementary Information
ijms-14-14996-s001.pdfAcknowledgments
Conflict of Interest
References
- Van Poecke, R.M.P. Arabidopsis-insect interactions. Arabidopsis B 2007, 5, e0107. [Google Scholar]
- Glazebrook, J. Contrasting mechanisms of defense against biotrophic and necrotrophic pathogens. Annu. Rev. Phytopathol 2005, 43, 205–227. [Google Scholar]
- Koornneef, A.; Pieterse, C.M.J. Cross talk in defense signaling. Plant Physiol 2008, 146, 839–844. [Google Scholar]
- Pieterse, C.M.J.; Dicke, M. Plant interactions with microbes and insects: from molecular mechanisms to ecology. Trends Plant Sci 2007, 12, 564–569. [Google Scholar]
- Clarke, D.B. Glucosinolates, structures and analysis in food. Anal. Methods 2010, 2, 310–325. [Google Scholar]
- Fahey, J.W.; Zhang, Y.S.; Talalay, P. Broccoli sprouts: an exceptionally rich source of inducers of enzymes that protect against chemical carcinogens. Proc. Nat. Acad. Sci. USA 1997, 94, 10367–10372. [Google Scholar]
- Block, G.; Patterson, B.; Subar, A. Fruit, vegetables, and cancer prevention: a review of the epidemiological evidence. Nutr. Cancer 1992, 18, 1–29. [Google Scholar]
- Wittstock, U.; Burow, M. Glucosinolate breakdown in arabidopsis: mechanism, regulation and biological significance. Arabidopsis B. 2010. [Google Scholar] [CrossRef]
- Halkier, B.A.; Gershenzon, J. Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol 2006, 57, 303–333. [Google Scholar]
- Sonderby, I.E.; Geu-Flores, F.; Halkier, B.A. Biosynthesis of glucosinolates—gene discovery and beyond. Trends Plant Sci 2010, 15, 283–290. [Google Scholar]
- Reichelt, M.; Brown, P.D.; Schneider, B.; Oldham, N.J.; Stauber, E.; Tokuhisa, J.; Kliebenstein, D.J.; Mitchell-Olds, T.; Gershenzon, J. Benzoic acid glucosinolate esters and other glucosinolates from Arabidopsis thaliana. Phytochemistry 2002, 59, 663–671. [Google Scholar]
- Rohr, F.; Ulrichs, C.; Schreiner, M.; Nguyen, C.N.; Mewis, I. Impact of hydroxylated and non-hydroxylated aliphatic glucosinolates in arabidopsis thaliana crosses on plant resistance against a generalist and a specialist herbivore. Chemoecology 2011, 21, 171–180. [Google Scholar]
- Mewis, I.; Appel, H.M.; Hom, A.; Raina, R.; Schultz, J.C. Major signaling pathways modulate arabidopsis glucosinolate accumulation and response to both phloem-feeding and chewing Insects. Plant Physiol 2005, 138, 1149–1162. [Google Scholar]
- Beran, F. Host preference and aggregation behavior of the striped flea beetle, phyllotreta striolata. In Ph.D. Thesis; the Humboldt-Universität: Berlin, Germany, 2011. [Google Scholar]
- Kim, J.H.; Jander, G. Myzus persicae (Green Peach Aphid) Feeding on Arabidopsis Induces the Formation of a Deterrent Indole Glucosinolate. Plant J 2007, 49, 1008–1019. [Google Scholar]
- De Vos, M.; Kriksunov, K.L.; Jander, G. Indole-3-acetonitrile Production from Indole Glucosinolates Deters Oviposition by Pieris rapae. Plant Physiol 2008, 146, 916–926. [Google Scholar]
- Brown, P.D.; Tokuhisa, J.G.; Reichelt, M.; Gershenzon, J. Variation of Glucosinolate Accumulation among Different Organs and Developmental Stages of Arabidopsis thaliana. Phytochemistry 2003, 62, 471–481. [Google Scholar]
- Gimsing, A.L.; Kirkegaard, J.A. Glucosinolates and biofumigation: Fate of glucosinolates and their hydrolysis products in soil. Phytochem. Rev 2009, 8, 299–310. [Google Scholar]
- Baasanjav-Gerber, C.; Monien, B.H.; Mewis, I.; Schreiner, M.; Barillari, J.; Iori, R.; Glatt, H. Identification of glucosinolate congeners able to form dna adducts and to induce mutations upon activation by myrosinase. Mol. Nutr. Food Res 2011, 55, 783–792. [Google Scholar]
- Glatt, H.; Baasanjav-Gerber, C.; Schumacher, F.; Monien, B.H.; Schreiner, M.; Frank, H.; Seidel, A.; Engst, W. 1-Methoxy-3-indolylmethyl glucosinolate; a potent genotoxicant in bacterial and mammalian cells: mechanisms of bioactivation. Chem.-Biol. Interact 2011, 192, 81–86. [Google Scholar]
- Pfalz, M.; Mikkelsen, M.D.; Bednarek, P.; Olsen, C.E.; Halkier, B.A.; Kroymann, J. Metabolic engineering in Nicotiana benthamiana Reveals key enzyme functions in arabidopsis indole glucosinolate modification. Plant Cell 2011, 23, 716–729. [Google Scholar]
- Mikkelsen, M.D.; Petersen, B.L.; Glawischnig, E.; Jensen, A.B.; Andreasson, E.; Halkier, B.A. Modulation of CYP79 genes and glucosinolate profiles in Arabidopsis by defense signaling pathways. Plant Physiol 2003, 131, 298–308. [Google Scholar]
- Bodnaryk, R.P. Potent effect of jasmonates on indole glucosinolates in oilseed rape and mustard. Phytochemistry 1994, 35, 301–305. [Google Scholar]
- Kiddle, G.A.; Doughty, K.J.; Wallsgrove, R.M. Salicylic acid-induced accumulation of glucosinolates in oilseed rape (Brassica napus L.) leaves. J. Exp. Bot 1994, 45, 1343–1346. [Google Scholar]
- Doughty, K.J.; Kiddle, G.A.; Pye, B.J.; Wallsgrove, R.M.; Pickett, J.A. selective induction of glucosinolates in oilseed rape leaves by methyl jasmonate. Phytochemistry 1995, 38, 347–350. [Google Scholar]
- Smetanska, I.; Krumbein, A.; Schreiner, M.; Knorr, D. Influence of salicylic acid and methyl jasmonate on glucosinolate levels in turnip. J. Hortic. Sci. Biotech 2007, 82, 690–694. [Google Scholar]
- Schreiner, M.; Krumbein, A.; Knorr, D.; Smetanska, I. Enhanced glucosinolates in root exudates of brassica rapa ssp rapa mediated by salicylic acid and methyl jasmonate. J. Agric. Food Chem 2011, 59, 1400–1405. [Google Scholar]
- Perez-Balibrea, S.; Moreno, D.A.; Garcia-Viguera, C. Improving the phytochemical composition of broccoli sprouts by elicitation. Food Chem 2011, 129, 35–44. [Google Scholar]
- Sun, B.; Yan, H.; Zhang, F.; Wang, Q. Effects of plant hormones on main health-promoting compounds and antioxidant capacity of chinese kale. Food Res. Int 2012, 48, 359–366. [Google Scholar]
- Smolen, G.; Bender, J. Arabidopsis cytochrome P450 CYP83B1 mutations activate the tryptophan biosynthetic pathway. Genetics 2002, 160, 323–332. [Google Scholar]
- Dombrecht, B.; Xue, G.P.; Sprague, S.J.; Kirkegaard, J.A.; Ross, J.J.; Reid, J.B.; Fitt, G.P.; Sewelam, N.; Schenk, P.M.; Manners, J.M.; et al. MYC2 differentially modulates diverse jasmonate-dependent functions in Arabidopsis. Plant Cell 2007, 19, 2225–2245. [Google Scholar]
- Doares, S.H.; Narvaezvasquez, J.; Conconi, A.; Ryan, C.A. Salicylic acid inhibits synthesis of proteinase-inhibitors in tomato leaves induced by systemin and jasmonic acid. Plant Physiol 1995, 108, 1741–1746. [Google Scholar]
- Pena-Cortes, H.; Albrecht, T.; Prat, S.; Weiler, E.W.; Willmitzer, L. Aspirin prevents wound-induced gene-expression in tomato leaves by blocking jasmonic acid biosynthesis. Planta 1993, 191, 123–128. [Google Scholar]
- Wiesner, M.; Zrenner, R.; Krumbein, A.; Glatt, H.; Schreiner, M. Genotypic variation of the glucosinolate profile in pak choi (Brassica rapa ssp. chinensis). J. Agric. Food Chem 2013, 61, 1943–1953. [Google Scholar]
- Wang, X.; Wang, H.; Wang, J.; Sun, R.; Wu, J.; Liu, S.; Bai, Y.; Mun, J.H.; Bancroft, I.; Cheng, F.; et al. The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet 2011, 43, 1035–1039. [Google Scholar]
- Van Dam, N.M.; Oomen, M.W.A.T. Root and shoot jasmonic acid applications differentially affect leaf chemistry and herbivore growth. Plant Signal. Behav 2008, 3, 91–98. [Google Scholar]
- Zang, Y.X.; Kim, H.U.; Kim, J.A.; Lim, M.H.; Jin, M.; Lee, S.C.; Kwon, S.J.; Lee, S.I.; Hong, J.K.; Park, T.H.; et al. Genome-wide identification of glucosinolate synthesis genes in Brassica rapa. FEBS J 2009, 276, 3559–3574. [Google Scholar]
- Textor, S.; Gershenzon, J. Herbivore induction of the glucosinolate-myrosinase defense system: major trends, biochemical bases and ecological significance. Phytochem. Rev 2009, 8, 149–170. [Google Scholar]
- Sasaki-Sekimoto, Y.; Taki, N.; Obayashi, T.; Aono, M.; Matsumoto, F.; Sakurai, N.; Suzuki, H.; Hirai, M.Y.; Noji, M.; Saito, K.; et al. Coordinated activation of metabolic pathways for antioxidants and defence compounds by jasmonates and their roles in stress tolerance in Arabidopsis. Plant J 2005, 44, 653–668. [Google Scholar]
- Loivamäki, M.; Holopainen, J.K.; Nerg, A.M. Chemical changes induced by methyl jasmonate in oilseed rape grown in the laboratory and in the field. J. Agric. Food Chem 2004, 52, 7607–7613. [Google Scholar]
- Mewis, I.; Schreiner, M.; Nguyen, C.N.; Krumbein, A.; Ulrichs, C.; Lohse, M.; Zrenner, R. UV-B irradiation changes specifically the secondary metabolite profile in broccoli sprouts: Induced signaling overlaps with defense response to biotic stressors. Plant Cell Physiol 2012, 53, 1546–1560. [Google Scholar]
- Gigolashvili, T.; Berger, B.; Mock, H.P.; Muller, C.; Weisshaar, B.; Fluegge, U.I. The transcription factor hig1/myb51 regulates indolic glucosinolate biosynthesis in Arabidopsis thaliana. Plant J 2007, 50, 886–901. [Google Scholar]
- Agerbirk, N.; de Vos, M.; Kim, J.H.; Jander, G. Indole glucosinolate breakdown and its biological effects. Phytochem. Rev 2009, 8, 101–120. [Google Scholar]
- Hanley, A.B.; Parsley, K.R. Identification of 1-methoxyindolyl-3-methyl isothiocyanate as an indole glucosinolate breakdown product. Phytochemistry 1990, 29, 769–771. [Google Scholar]
- Malitsky, S.; Blum, E.; Less, H.; Venger, I.; Elbaz, M.; Morin, S.; Eshed, Y.; Aharoni, A. The transcript and metabolite networks affected by the two clades of arabidopsis glucosinolate biosynthesis regulators. Plant Physiol 2008, 148, 2021–2049. [Google Scholar]
- Elbaz, M.; Halon, E.; Malka, O.; Malitsky, S.; Blum, E.; Aharoni, A.; Morin, S. Asymmetric adaptation to indolic and aliphatic glucosinolates in the b and q sibling species of Bemisia tabaci (Hemiptera: Aleyrodidae). Mol. Ecol 2012, 21, 4533–4546. [Google Scholar]
- Clay, N.K.; Adio, A.M.; Denoux, C.; Jander, G.; Ausubel, F.M. Glucosinolate metabolites required for an arabidopsis innate immune response. Science 2009, 323, 95–101. [Google Scholar]
- Bednarek, P.; Pislewska-Bednarek, M.; Svatos, A.; Schneider, B.; Doubsky, J.; Mansurova, M.; Humphry, M.; Consonni, C.; Panstruga, R.; Sanchez-Vallet, A.; et al. Glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 2009, 323, 101–106. [Google Scholar]
- Ruijter, J.; Ramakers, C.; Hoogaars, W.; Karlen, Y.; Bakker, O.; van den Hoff, M.; Moorman, A. Amplification efficiency: linking baseline and bias in the analysis of quantitative pcr data. Nucleic Acids Res. 2009, 37. [Google Scholar] [CrossRef]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative pcr and the 2(t)(-delta delta c) method. Methods 2001, 25, 402–408. [Google Scholar]
- Hanschen, F.S.; Bauer, A.; Mewis, I.; Keil, C.; Schreiner, M.; Rohn, S.; Kroh, L.W. Thermally induced degradation of aliphatic glucosinolates: identification of intermediary breakdown products and proposed degradation pathways. J. Agric. Food Chem 2012, 60, 9890–9899. [Google Scholar]
- STATISTICA, version 10; software for anlysing data; StatSoft Inc.: Tulsa, OK, USA, 2011.







| Gene function (Gene name) | Oligonucleotide abbreviation | Sequence | Accession (Gene abbr.) |
|---|---|---|---|
| Actin2 (ACT2) | Br-Af | ACGTGGACATCAGGAAGGAC | AC189447 |
| Br-Br | CTTGGTGCAAGTGCTGTGAT | (BrACT2) | |
| Transcription factor Altered Tryptophan Regulation 1 (ATR1/MYB34) | Br-Myb34f1 | GAAGGAATAAAGAAAGGAGCC | FJ584293 |
| Br-Myb34r1 | GCATCTCTTCAATCCAGCTTTT | (BrMYB34_1) | |
| Br-Myb34f2 | AGAAGGAATAAAGAAAGGAGCT | FJ584294 | |
| Br-Myb34r1 | (BrMYB34_2) | ||
| Br-Myb34f2 | FJ584295 | ||
| Br-Myb34r2 | CATCTCTTCAATCCAGCTTTC | (BrMYB34_3) | |
| Transcription factor High Indolic Glucosinolate 1 (HIG1/MYB51) | Br-Myb51f1 | ACCGATAACGAAATCAAGAACTA | FJ584296 |
| Br-Myb51r1 | CTCACAAGAACATATCAGAAAATT | (BrMYB51_1) | |
| Br-Myb51f2 | ACCGATAACGAAATCAAGAACCA | FJ584297 | |
| Br-Myb51r2 | TCCGACAAATCAGAAAACCTCC | (BrMYB51_2) | |
| Br-Myb51f3 | FJ584299 | ||
| Br-Myb51r2 | ACCGATAACGAAATCAAGAACCT | (BrMYB51_3) | |
| Br-Myb51f1 | FJ584298 | ||
| Br-Myb51r3 | TCCGAAACCAGCAAATCAGAAA | (BrMYB51_4) | |
| Transcription factor High Indolic Glucosinolate 2 (HIG2/MYB122) | Br-Myb122f1 | TGGATGAATCTTCTTCAGACAAT | FJ584300 |
| Br-Myb122r1 | ATGTCGCTTAATGATAGCCACC | (BrMYB122_1) | |
| Br-Myb122f2 | TGGATGAATCTTGTTTGGAGAAA | Bra008131 | |
| Br-Myb122r2 | ACTATGTGTAGTGATAGTCGTG | (BrMYB122_2) | |
| Cytochrom P450 monooxygenase family 79B (CYP79B) | Br-Cyp79B2f3 | GTTCTCTGAAAACACTGCAGCG | FJ376045 |
| Br-Cyp79B2r3 | TGCTTTTTGTATATCTGATTATCTAC | (BrCYP79B2_1) | |
| Br-Cyp79B2f4 | GTTCTCTGAAAACACCGCACCT | FJ376047 | |
| Br-Cyp79B2r4 | CTCCTTTTGTATCTCTGATTATCTA | (BrCYP79B2_2) | |
| Br-Cyp79B3f | TTCTCGGAGAAAACCAAAACC | FJ376047 | |
| Br-Cyp79B3r | TGCGTTTTGTGTATCGGACTA | (BrCYP79B3) | |
| Sulfotransferase 16 (SOT16) | Br-SOT16f2 | TTCAAGACGGCAAGAACCAG | FJ376059 |
| Br-SOT16r2 | GGGTCAGCAGCTAGCGAG | (BrSOT16) | |
| Cytochrom P450 monooxygenase family 81F (CYP81F) | Br-Cyp81F1f | TCCCTCGCACGCCGACG | KBrB006J12.9 |
| Br-Cyp81F1r | AGGATGCGGCAGCGAGTTA | (BrCYP81F1) | |
| Br-Cyp81F2f | TCTCCTTCTGAAGATCTCAAAA | KBrB027E01.6 | |
| Br-Cyp81F2r | AGAAAAAGAAGCAGCGAACAC | (BrCYP81F2) | |
| Br-Cyp81F3f1 | GCCGAGATCACCGATGGAA | KBrB006J12.6 | |
| Br-Cyp81F3r1 | GCGGAGGAGAAGACGTTCA | (BrCYP81F3_1) | |
| Br-Cyp81F3f2 | GCCAAGATCGACGACAGAC | KBrH064I20.2 | |
| Br-Cyp81F3r2 | TCGGAGAAGGAGGAGAAGAC | (BrCYP81F3_2) | |
| Br-Cyp81F4f1 | TTAACGGAAGAGGACATCAAAG | KBrB006J12.7 | |
| Br-Cyp81F4r1 | AAAGAGGGGAAGGAGACAAAGA | (BrCYP81F4_1) | |
| Br-Cyp81F4f2 | TTAACAGTAGAGGACATCAAGA | KBrH064I20.1 | |
| Br-Cyp81F4r2 | TGGAGGAGAAGGAGAAAAGGA | (BrCYP81F4_2) | |
| O-methyltransferase family protein (OMT) | Br-OMTf1 | GGCTGTACCGGAGAGACGA | Bra017700 |
| Br-OMTr1 | GCCGTTCTCATCAAGTGGGTG | (BrOMT1_1) | |
| Br-OMTf2 | GGCTGTACCGGAGAGACCC | Bra017699 | |
| Br-OMTr2 | AACGTTTTCATCAAGTGGGTCT | (BrOMT1_2) | |
| Control | MeJA treated plants | |
|---|---|---|
| Glucosinolate level (μmol g−1 dry weight) | ||
| Aliphatic glucosinolates | 2.80 ±1.24 | 6.20 ± 2.15 |
| Indol-3-ylmethyl | 0.26 ± 0.07 | 4.24 ± 0.73 * |
| 4-Hydroxy-indol-3-ylmethyl | 0.06 ± 0.02 | 0.21 ± 0.05 * |
| 4-Methoxy-indol-3-ylmethyl | 0.08 ± 0.01 | 0.11 ± 0.03 |
| 1-Methoxy-indol-3-ylmethyl | 0.07 ± 0.04 | 5.84 ± 0.66 * |
| Breakdown products (μmol g−1 fresh weight) | ||
| Aliphatic isothiocyanates, nitriles | 0.251 ± 0.202 | 0.199 ± 0.120 |
| Indole-acetonitrile | 0.017 ± 0.018 | 0.246 ± 0.094 * |
| 1-Methoxy-indole-acetonitrile | 0.001 ± 0.003 | 0.079 ± 0.031 * |
| 1-Methoxy-indole-3-carbinole | 0.000 ± 0.000 | 0.057 ± 0.024 * |
© 2013 by the authors; licensee MDPI, Basel, Switzerland This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/3.0/).
Share and Cite
Wiesner, M.; Hanschen, F.S.; Schreiner, M.; Glatt, H.; Zrenner, R. Induced Production of 1-Methoxy-indol-3-ylmethyl Glucosinolate by Jasmonic Acid and Methyl Jasmonate in Sprouts and Leaves of Pak Choi (Brassica rapa ssp. chinensis). Int. J. Mol. Sci. 2013, 14, 14996-15016. https://doi.org/10.3390/ijms140714996
Wiesner M, Hanschen FS, Schreiner M, Glatt H, Zrenner R. Induced Production of 1-Methoxy-indol-3-ylmethyl Glucosinolate by Jasmonic Acid and Methyl Jasmonate in Sprouts and Leaves of Pak Choi (Brassica rapa ssp. chinensis). International Journal of Molecular Sciences. 2013; 14(7):14996-15016. https://doi.org/10.3390/ijms140714996
Chicago/Turabian StyleWiesner, Melanie, Franziska S. Hanschen, Monika Schreiner, Hansruedi Glatt, and Rita Zrenner. 2013. "Induced Production of 1-Methoxy-indol-3-ylmethyl Glucosinolate by Jasmonic Acid and Methyl Jasmonate in Sprouts and Leaves of Pak Choi (Brassica rapa ssp. chinensis)" International Journal of Molecular Sciences 14, no. 7: 14996-15016. https://doi.org/10.3390/ijms140714996
APA StyleWiesner, M., Hanschen, F. S., Schreiner, M., Glatt, H., & Zrenner, R. (2013). Induced Production of 1-Methoxy-indol-3-ylmethyl Glucosinolate by Jasmonic Acid and Methyl Jasmonate in Sprouts and Leaves of Pak Choi (Brassica rapa ssp. chinensis). International Journal of Molecular Sciences, 14(7), 14996-15016. https://doi.org/10.3390/ijms140714996