FT-IR Microspectrometry Reveals the Variation of Membrane Polarizability due to Epigenomic Effect on Epithelial Ovarian Cancer
Abstract
:1. Introduction


2. Results and Discussion
2.1. 5-Aza-2'-deoxycytidine (5-aza) Weakens Methylation of Deoxycytidine Monophosphate (dCMP) and Enhances Protein Expression in CP70

| Sample | Treatment | Retention Time (min) | Ratio of Methylation a | |
|---|---|---|---|---|
| dCMP | mdCMP | |||
| Standard | 3.6 ± 0.1 | 3.9 ± 0.1 | ||
| IOSE | None | 3.6 ± 0.1 | 4.1 ± 0.1 | 4.3% ± 1.7 |
| 5-Aza | 3.6 ± 0.1 | 3.9 ± 0.1 | 4.2% ± 0.8 | |
| A2780 | None | 3.5 ± 0.1 | 4.0 ± 0.1 | 5.4% ± 2.4 |
| 5-Aza | 3.6 ± 0.1 | 4.1 ± 0.1 | 1.3% ± 0.1 | |
| CP70 | None | 3.6 ± 0.1 | 4.0 ± 0.1 | 7.7% ± 2.0 |
| 5-Aza | 3.6 ± 0.1 | 4.1 ± 0.1 | 4.8% ± 0.6 | |
| Cell Line | No Treatment | 5-Aza-Treatment |
|---|---|---|
| IOSE | 13.7 ± 1.2 | 15.1 ± 2.1 |
| A2780 | 11.9 ± 1.0 | 15.6 ± 3.4 |
| CP70 | 11.3 ± 1.4 | 17.8 ± 2.0 |
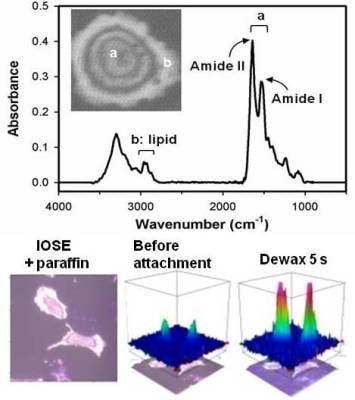
2.2. Distinguishing Ovarian Normal and Cancer Cells with Paraffin-Adsorption Kinetics

3. Experimental Section
3.1. Cell Culture and Drug Treatments
3.2. Quantitative Real-Time Methylation Specific PCR (qMSP)
3.3. DNA Analysis with Reverse-Phase HPLC Measurement
3.4. FT-IR Spectromicroscopy of Ovarian Cells
3.5. Wax-Adsorption Kinetic Study with FT-IR Microspectrometry
3.6. Analysis of Spectra Data
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Jemal, A.; Murray, T.; Samuels, A.; Ghafoor, A.; Ward, E.; Thun, M.J. Cancer statistics, 2003. CA Cancer J. Clin. 2003, 53, 5–26. [Google Scholar]
- Holschneider, C.H.; Berek, J.S. Ovarian cancer: Epidemiology, biology, and prognostic factors. Semin. Surg. Oncol. 2000, 19, 3–10. [Google Scholar]
- Yin, B.W.; Lloyd, K.O. Molecular cloning of the CA125 ovarian cancer antigen: Identification as a new mucin, MUC16. J. Biol. Chem. 2001, 276, 27371–27375. [Google Scholar]
- Yin, B.W.; Dnistrian, A.; Lloyd, K.O. Ovarian cancer antigen CA125 is encoded by the MUC16 mucin gene. Int. J. Cancer 2002, 98, 737–740. [Google Scholar]
- Bast, R.C., Jr.; Xu, F.J.; Yu, Y.H.; Barnhill, S.; Zhang, Z.; Mills, G.B. CA125: The past and the future. Int. J. Biol. Mark. 1998, 13, 179–187. [Google Scholar]
- Bird, A. DNA methylation patterns and epigenetic memory. Genes Dev. 2002, 16, 6–21. [Google Scholar]
- Christman, J.K. 5-Azacytidine and 5-aza-2'-deoxycytidine as inhibitors of DNA methylation: Mechanistic studies and their implications for cancer therapy. Oncogene 2002, 21, 5483–5495. [Google Scholar]
- Auersperg, N.; Pan, J.; Grove, B.D.; Peterson, T.; Fisher, J.; Maines-Bandiera, S.; Somasiri, A.; Roskelley, C.D. E-cadherin induces mesenchymal-to-epithelial transition in human ovarian surface epithelium. Proc. Natl. Acad. Sci. USA 1999, 96, 6249–6254. [Google Scholar]
- Ong, A.; Maines-Bandiera, S.L.; Roskelley, C.D.; Auersperg, N. An ovarian adenocarcinoma line derived from SV40/E-cadherin-transfected normal human ovarian surface epithelium. Int. J. Cancer 2000, 85, 430–437. [Google Scholar]
- Maines-Bandiera, S.L.; Kruk, P.A.; Auersperg, N. Simian virus 40-transformed human ovarian surface epithelial cells escape normal growth controls but retain morphogenetic responses to extracellular matrix. Am. J. Obstet. Gynecol. 1992, 167, 729–735. [Google Scholar]
- Cheng, P.; Schmutte, C.; Cofer, K.F.; Felix, J.C.; Yu, M.C.; Dubeau, L. Alterations in DNA methylation are early, but not initial, events in ovarian tumorigenesis. Br. J. Cancer 1997, 75, 396–402. [Google Scholar]
- Ehrlich, M. DNA methylation in cancer: Too much, but also too little. Oncogene 2002, 21, 5400–5413. [Google Scholar]
- Gama-Sosa, M.A.; Slagel, V.A.; Trewyn, R.W.; Oxenhandler, R.; Kuo, K.C.; Gehrke, C.W.; Ehrlich, M. The 5-methylcytosine content of DNA from human tumors. Nucleic Acids Res. 1983, 11, 6883–6894. [Google Scholar]
- Muller, H.M.; Widschwendter, M. Methylated DNA as a possible screening marker for neoplastic disease in several body fluids. Expert Rev. Mol. Diagn. 2003, 3, 443–458. [Google Scholar]
- Strathdee, G.; MacKean, M.J.; Illand, M.; Brown, R. A role for methylation of the hMLH1 promoter in loss of hMLH1 expression and drug resistance in ovarian cancer. Oncogene 1999, 18, 2335–2341. [Google Scholar]
- Plumb, J.A.; Steele, N.; Finn, P.W.; Brown, R. Epigenetic approaches to cancer therapy. Biochem. Soc. Trans. 2004, 32, 1095–1097. [Google Scholar]
- Brenner, T.L.; Adams, V.R. First MAb approved for treatment of metastatic breast cancer. J. Am. Pharm. Assoc. 1999, 39, 236–238. [Google Scholar]
- Abicht, A.; Lochmuller, H. Technology evaluation: Edrecolomab, Centocor Inc. Curr. Opin. Mol. Ther. 2000, 2, 593–600. [Google Scholar]
- Ranson, M.; Mansoor, W.; Jayson, G. ZD1839 (IRESSA): A selective EGFR-TK inhibitor. Expert Rev. Anticancer Ther. 2002, 2, 161–168. [Google Scholar]
- Yano, K.; Ohoshima, S.; Shimizu, Y.; Moriguchi, T.; Katayama, H. Evaluation of glycogen level in human lung carcinoma tissues by an infrared spectroscopic method. Cancer Lett. 1996, 110, 29–34. [Google Scholar]
- Hammody, Z.; Argov, S.; Sahu, R.K.; Cagnano, E.; Moreh, R.; Mordechai, S. Distinction of malignant melanoma and epidermis using IR micro-spectroscopy and statistical methods. Analyst 2008, 133, 372–378. [Google Scholar]
- Kelly, J.G.; Singh, M.N.; Stringfellow, H.F.; Walsh, M.J.; Nicholson, J.M.; Bahrami, F.; Ashton, K.M.; Pitt, M.A.; Martin-Hirsch, P.L.; Martin, F.L. Derivation of a subtype-specific biochemical signature of endometrial carcinoma using synchrotron-based Fourier-transform infrared microspectroscopy. Cancer Lett. 2009, 274, 208–217. [Google Scholar]
- Wood, B.R.; Chiriboga, L.; Yee, H.; Quinn, M.A.; McNaughton, D.; Diem, M. Fourier transform infrared (FTIR) spectral mapping of the cervical transformation zone, and dysplastic squamous epithelium. Gynecol. Oncol. 2004, 93, 59–68. [Google Scholar]
- Eikje, N.S.; Aizawa, K.; Ozaki, Y. Vibrational spectroscopy for molecular characterisation and diagnosis of benign, premalignant and malignant skin tumours. Biotechnol. Annu. Rev. 2005, 11, 191–225. [Google Scholar]
- Krafft, C.; Shapoval, L.; Sobottka, S.B.; Geiger, K.D.; Schackert, G.; Salzer, R. Identification of primary tumors of brain metastases by SIMCA classification of IR spectroscopic images. Biochim. Biophys. Acta 2006, 1758, 883–891. [Google Scholar]
- Holman, H.Y.; Martin, M.C.; Blakely, E.A.; Bjornstad, K.; McKinney, W.R. IR spectroscopic characteristics of cell cycle and cell death probed by synchrotron radiation based Fourier transform IR spectromicroscopy. Biopolymers 2000, 57, 329–335. [Google Scholar]
- Chiu, L.F.; Huang, P.Y.; Chiang, W.F.; Wong, T.Y.; Lin, S.H.; Lee, Y.C.; Shieh, D.B. Oral cancer diagnostics based on infrared spectral markers and wax physisorption kinetics. Anal. Bioanal. Chem. 2013, 405, 1995–2007. [Google Scholar]
- Chan, M.W.; Wei, S.H.; Wen, P.; Wang, Z.; Matei, D.E.; Liu, J.C.; Liyanarachchi, S.; Brown, R.; Nephew, K.P.; Yan, P.S.; et al. Hypermethylation of 18S and 28S ribosomal DNAs predicts progression-free survival in patients with ovarian cancer. Clin. Cancer Res. 2005, 11, 7376–7383. [Google Scholar]
- Widschwendter, M.; Jiang, G.; Woods, C.; Muller, H.M.; Fiegl, H.; Goebel, G.; Marth, C.; Muller-Holzner, E.; Zeimet, A.G.; Laird, P.W.; et al. DNA hypomethylation and ovarian cancer biology. Cancer Res. 2004, 64, 4472–4480. [Google Scholar]
- Ramsahoye, B.H. Measurement of genome wide DNA methylation by reversed-phase high-performance liquid chromatography. Methods 2002, 27, 156–161. [Google Scholar]
- McGhee, J.D.; Ginder, G.D. Specific DNA methylation sites in the vicinity of the chicken β-globin genes. Nature 1979, 280, 419–420. [Google Scholar]
- Kampen, K.R. Membrane proteins: The key players of a cancer cell. J. Membr. Biol. 2011, 242, 69–74. [Google Scholar]
- Chou, J.L.; Su, H.Y.; Chen, L.Y.; Liao, Y.P.; Hartman-Frey, C.; Lai, Y.H.; Yang, H.W.; Deatherage, D.E.; Kuo, C.T.; Huang, Y.W.; et al. Promoter hypermethylation of FBXO32, a novel TGF-β/SMAD4 target gene and tumor suppressor, is associated with poor prognosis in human ovarian cancer. Lab. Investig. 2010, 90, 414–425. [Google Scholar]
© 2014 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hsu, M.M.H.; Huang, P.-Y.; Lee, Y.-C.; Fang, Y.-C.; Chan, M.W.Y.; Lee, C.-I. FT-IR Microspectrometry Reveals the Variation of Membrane Polarizability due to Epigenomic Effect on Epithelial Ovarian Cancer. Int. J. Mol. Sci. 2014, 15, 17963-17973. https://doi.org/10.3390/ijms151017963
Hsu MMH, Huang P-Y, Lee Y-C, Fang Y-C, Chan MWY, Lee C-I. FT-IR Microspectrometry Reveals the Variation of Membrane Polarizability due to Epigenomic Effect on Epithelial Ovarian Cancer. International Journal of Molecular Sciences. 2014; 15(10):17963-17973. https://doi.org/10.3390/ijms151017963
Chicago/Turabian StyleHsu, Morris M. H., Pei-Yu Huang, Yao-Chang Lee, Yuang-Chuen Fang, Michael W. Y. Chan, and Cheng-I Lee. 2014. "FT-IR Microspectrometry Reveals the Variation of Membrane Polarizability due to Epigenomic Effect on Epithelial Ovarian Cancer" International Journal of Molecular Sciences 15, no. 10: 17963-17973. https://doi.org/10.3390/ijms151017963
APA StyleHsu, M. M. H., Huang, P. -Y., Lee, Y. -C., Fang, Y. -C., Chan, M. W. Y., & Lee, C. -I. (2014). FT-IR Microspectrometry Reveals the Variation of Membrane Polarizability due to Epigenomic Effect on Epithelial Ovarian Cancer. International Journal of Molecular Sciences, 15(10), 17963-17973. https://doi.org/10.3390/ijms151017963