Graphene Functionalized with Arginine Decreases the Development of Glioblastoma Multiforme Tumor in a Gene-Dependent Manner
Abstract
:1. Introduction
2. Results
2.1. Characterization of GO (Graphene Oxide) and rGO (Reduced Graphene Oxide)


2.2. Experiments with Fibroblast and Glioblastoma Cells
Mortality, Viability and Morphology of Cells




2.3. Experiments with Tumor Tissue
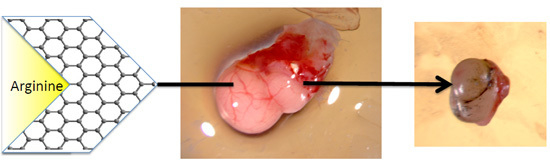
2.3.1. The Volume, Weight and Morphology of GBM Tumors
| Parameter | Group | ANOVA | ||||
|---|---|---|---|---|---|---|
| Control | rGO | rGO + Arg | rGO + Pro | SE | p-Value | |
| Weight [mg] | 0.0981 a | 0.0637 b | 0.0666 b | 0.0598 b | 0.01551 | 0.0317 |
| Volume [mm3] | 114.2 a | 45.32 b | 74.51 b | 64.12 b | 18.481 | 0.0049 |

2.3.2. Histology of Tumors

2.3.3. Gene Expression in Tumors
| Genes | Group | ANOVA | ||||
|---|---|---|---|---|---|---|
| Control | rGO | rGO + Arg | rGO + Pro | SE | p-Value | |
| TP53 | 0.345 a | 1.310 b | 0.965 c | 0.604 a | 0.0999 | 0.0001 |
| MDM2 | 0.845 a | 3.053 b | 1.919 a | 1.564 a | 0.3544 | 0.0065 |
| MDM2/TP53 | 2.45 a | 2.33 a | 1.99 b | 2.59 a | 0.288 | 0.0032 |
| COX6 | 0.323 a | 0.950 c | 1.082 c | 0.714 b | 0.0777 | 0.0001 |
| NQO1 | 0.380 a | 3.781 b | 10.845 c | 5.012 b | 0.2208 | 0.0000 |
| CASP3 | 0.428 a | 0.920 b | 1.052 b | 0.780 a,b | 0.1724 | 0.0115 |
| FGF2 | 1.059 a | 0.671 b | 0.432 b | 0.741 b | 0.0956 | 0.0045 |
| VEGF | 0.528 | 0.438 | 0.287 | 0.526 | 0.1723 | 0.1156 |
3. Discussion
4. Experimental Section
4.1. Preparation of Graphene Complexes
4.2. Cell Culture
4.3. Cell Morphology
4.4. Cell Mortality
4.5. Cell Viability
4.6. Culture of GMB on a Chorioallantoic Membrane
4.7. Tumor Volume and Histology
4.8. Gene Expression at the mRNA Level
| Gene | Forward Primer (5′–3′) | Reverse Primer (5′–3′) |
|---|---|---|
| TP53 | CCCAGCCAAAGAAGAAACCA | TTCCAAGGCCTCATTCAGCT |
| MDM2 | CAGGACATCTTATGGCCTGCTT | GGGCAGGGCTTATTCCTTTT |
| COX6 | TGAATCCGGGGTGCCTTTAG | CAGAGGGACTGGTACACACG |
| NQO1 | AGGCTGGTTTGAGCGTGTTC | TTGAATTCGGGCGTCTGCTG |
| CASP3 | ACATGGCGTGTCATAAAATACC | CACAAAGCGACTGGATGAAC |
| FGF2 | GGCACTGAAATGTGCAACAG | TCCAGGTCCAGTTTTTGGTC |
| VEGFA | TGAGGGCCTAGAATGTGTCC | TCTTTTGACCCTTCCCCTTT |
| ACTB | ACCCAGATCATGTTCGAGACCTT | TCACCGGAGTCCATCACGAT |
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Walker, C.; Baborie, A.; Crooks, D.; Wilkins, S.; Jenkinson, M.D. Biology, genetics and imaging of glial cell tumours. Br. J. Radiol. 2011, 84, 90–106. [Google Scholar] [CrossRef] [PubMed]
- DeAngelis, L.M. Medical progress: Brain tumors. N. Engl. J. Med. 2001, 344, 114–123. [Google Scholar] [CrossRef] [PubMed]
- Parson, D.W.; Jones, S.; Zhang, X.; Lin, J.C.; Leary, R.J.; Angenendt, P.; Mankoo, P.; Carter, H.; Siu, I.M.; Gallia, G.L.; et al. An Integrated Genomic Analysis of Human Glioblastoma Multiforme. Science 2008, 321, 1807–1812. [Google Scholar] [CrossRef] [PubMed]
- Shakur, S.F.; Bit-Ivan, E.; Watkin, W.G.; Merrell, R.T.; Farhat, H.I. Multifocal and multicentric glioblastoma with leptomeningeal gliomatosis: A case report and review of the literature. Case Rep. Med. 2013, 2013, 132679. [Google Scholar] [CrossRef] [PubMed]
- Kesari, S. Understanding glioblastoma tumor biology: The potential to improve current diagnosis and treatments. Semin. Oncol. 2011, 38, S2–S10. [Google Scholar] [CrossRef] [PubMed]
- Geim, A.K.; Novoselov, K.S. The rise of graphene. Nat. Mater. 2007, 6, 183–191. [Google Scholar] [CrossRef] [PubMed]
- Liu, Z.; Robinson, J.T.; Sun, X.; Dai, H. PEGylated nanographene oxide for delivery of water-insoluble cancer drugs. J. Am. Chem. Soc. 2008, 130, 10876–10877. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Cui, L.; Losic, D. Graphene and graphene oxide as new nanocarriers for drug delivery applications. Acta Biomater. 2013, 9, 9243–9257. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.M.; Liu, Z.; Welsher, K.; Robinson, J.T.; Godwin, A. Nano-graphene oxide for cellular imaging and drug delivery. Nano Res. 2008, 1, 203–212. [Google Scholar] [CrossRef] [PubMed]
- Jaworski, S.; Sawosz, E.; Grodzik, M.; Winnicke, A.; Prasek, M.; Wierzbicki, M.; Chwalibog, A. In vitro evaluation of the effects of graphene platelets on glioblastoma multiforme cells. Int. J. Nanomed. 2013, 8, 413–420. [Google Scholar]
- Fiorillo, M.; Verre, A.F.; Iliut, M.; Peiris-Pagés, M.; Ozsvari, B.; Gandara, R.; Cappello, A.R.; Sotgia, F.; Vijayaraghavan, A.; Lisanti, M.P. Graphene oxide selectively targets cancer stem cells, across multiple tumor types: Implications for non-toxic cancer treatment, via “differentiation-based nano-therapy”. Oncotarget 2015, 6, 3553–3562. [Google Scholar] [CrossRef] [PubMed]
- Konios, D.; Stylianakis, M.M.; Stratakis, E.; Kymakis, E. Dispersion behaviour of graphene oxide and reduced graphene oxide. J. Colloid Interface Sci. 2014, 430, 108–112. [Google Scholar] [CrossRef] [PubMed]
- Sawosz, E.; Jaworski, S.; Kutwin, M.; Hotowy, M.; Wierzbicki, M.; Grodzik, M.; Strojny, B.; Lipinska, L.; Chwalibog, A. Toxicity of pristine graphene in experiments in a chicken embryo model. Int. J. Nanomed. 2014, 9, 3913–3922. [Google Scholar]
- Jaworski, S.; Sawosz, E.; Kutwin, M.; Wierzbicki, M.; Hinzmann, M.; Grodzik, M.; Winnicka, A.; Lipinska, L.; Wlodyga, K.; Chwalibog, A. In vitro and in vivo effects of graphene oxide and reduced graphene oxide on glioblastoma multiforme. Int. J. Nanomed. 2015, 10, 1–12. [Google Scholar]
- Marchesan, S.; Prato, M. Under the lens: Carbon nanotube and protein interaction at the nanoscale. Chem. Commun. 2015, 51, 4347–4359. [Google Scholar] [CrossRef] [PubMed]
- Huges, Z.E.; Walsh, T.R. What makes a good graphene-binding peptide? Adsorption of amino acids and peptides at aqueous graphene interfaces. J. Mater. Chem. B 2015, 3, 3211–3221. [Google Scholar] [CrossRef]
- Kay, B.K.; Williamson, M.P.; Sudol, M. The importance of being proline: The interaction of proline-rich motifs in signaling proteins with their cognate domains. FASEB J. 2000, 14, 231–241. [Google Scholar] [PubMed]
- Krane, S.M. The importance of proline residues in the structures stability and susceptibility to proteolityc degradation of collagen. Amino Acids 2008, 35, 703–7010. [Google Scholar] [CrossRef] [PubMed]
- Betts, M.J.; Russell, R.B. Amino acids properties and consequences of substitution. In Bioinformatics for Geneticists, 2nd ed.; Barnes, M.R., Gray, I.C., Eds.; Wiley: Hoboken, NJ, USA, 2003; pp. 311–343. [Google Scholar]
- Hamasu, K.; Haraguchi, T.; Kabuki, Y.; Adachi, N.; Tomonaga, S.; Sato, H.; Denbow, D.M.; Furuse, M. l-Proline is a sedative regulator of acute stress in the brain of neonatal chicks. AMINO ACIDS 2009, 37, 377–382. [Google Scholar] [CrossRef] [PubMed]
- Phang, J.M.; Liu, W.; Hancock, C.; Christian, K.J. The proline regulatory axis and cancer. Front. Oncol. 2012, 2, 60. [Google Scholar] [CrossRef] [PubMed]
- Phang, J.M.; Donald, S.P.; Pandhare, J.; Liu, Y. The metabolism of proline, a stress substrate modulates carcinogenic pathway. Amino Acids 2008, 4, 726–745. [Google Scholar] [CrossRef] [PubMed]
- Bond, G.L.; Hu, W.; Levine, A.J. 2MDM2 is a central node in the p53 pathway: 12 years and counting. Curr. Cancer Drug Targets 2005, 6, 3–8. [Google Scholar] [CrossRef]
- Murphy, M.E. Regulation of IAP (Inhibitor of Apoptosis) Gene Expression by the p53 Tumor Expression Protein; Army Medical Research and Materiel Command Fort Detrick, Fox Chase Cancer Center: Philadelphia, PA, USA, 2005. [Google Scholar]
- Jansson, M.; Durant, S.T.; Cho, E.-C.; Sheahan, S.; Edelmann, M.; Kessler, B.; la Thangue, N.B. Arginine methylation regulates the p53 response. Nat. Cell Biol. 2008, 10, 1431–1439. [Google Scholar] [CrossRef] [PubMed]
- Jeong, B.-S.; Hu, W.; Belyi, V.; Rabadan, R.; Levine, A.J. Differential levels of transcription of p53-regulated genes by the arginine/proline polymorphism: p53 with arginine at codon 72 favors apoptosis. FASEB J. 2010, 24, 1347–1353. [Google Scholar] [CrossRef] [PubMed]
- Lin, W.-T.; Yang, S.-C.; Chen, K.-T.; Huang, C.-C.; Lee, N.-Y. Protective effects of l-arginine on pulmonary oxidative stress and antioxidant defenses during exhaustive exercise in rats. Acta Pharmacol. Sin. 2005, 26, 992–999. [Google Scholar] [CrossRef] [PubMed]
- Yeatman, T.J.; Risley, G.L.; Brunson, M.E. Depletion of dietary arginine inhibits growth of metastatic tumor. Arch. Surg. 1991, 126, 1376–1381. [Google Scholar] [PubMed]
- Roomi, M.W.; Ivanov, V.; Kalinovsky, T.; Niedzwiecki, A.; Rath, M. Antitumor effect of ascorbic acid, lysine, proline, arginine, and green tea extract on bladder cancer cell line T-24. Int. J. Urol. 2006, 13, 415–419. [Google Scholar] [CrossRef] [PubMed]
- Shu, X.; Liu, X.L.; Zhong, J.X.; Liu, J. l-Arginine enhance arginine deaminase induced human lymphoma cell growth inhibition through NF-κBp65 and p53 expression in vitro. Eur. Rev. Med. Pharmacol. Sci. 2014, 18, 2124–2131. [Google Scholar] [PubMed]
- Kurantowicz, N.; Sawosz, E.; Jaworski, S.; Kutwin, M.; Strojny, B.; Wierzbicki, M.; Szeliga, J.; Hotowy, A.; Lipińska, L.; Kozinski, R.; et al. Interaction of graphene family materials with Listeria monocytogenes and Salmonella enterica. Nanoscale Res. Lett. 2015, 10, 23. [Google Scholar] [CrossRef] [PubMed]
- Kumar, S.; Rai, S.B. Spectroscopic studies of l-arinine molecule. Indian J. Pure Appl. Phys. 2010, 48, 251–255. [Google Scholar]
- Shan, C.; Yang, H.; Han, D.; Zhang, Q.; Ivasak, A.; Niu, L. Water-soluble graphene covalently funcionalized by biocompatible poly-l-lysine. Langimuir 2009, 25, 12030–12033. [Google Scholar] [CrossRef] [PubMed]
- Singh, S.K.; Singh, M.K.; Kulkarni, P.P.; Sonkart, C.K.; Grácio, J.J.A. Amine-modified graphene: Thrombo-protective safer alternative to graphene oxide for biomedical applications. ACS Nano 2012, 6, 2731–2740. [Google Scholar] [CrossRef] [PubMed]
- Hinzmann, M.; Jaworski, S.; Kutwin, M.; Jagiełło, J.; Koziński, R.; Wierzbicki, M.; Grodzik, M.; Lipińska, L.; Sawosz, E.; Chwalibog, A. Nanoparticles containing allotropes of carbon have genotoxic effects on glioblastoma multiforme cells. Int. J. Nanomed. 2014, 9, 2409–2417. [Google Scholar]
- Liao, K.-H.; Lin, Y.-S.; Macosko, C.W.; Haynes, C.L. Cytotoxicity of graphene oxide and graphene in human erythrocytes and skin fibroblasts. ACS Appl. Mater. Interfaces 2011, 3, 2607–2615. [Google Scholar] [CrossRef] [PubMed]
- Saebra, A.B.; Paula, A.J.; de Lima, R.; Alves, O.L.; Duran, N. Nanotoxicity of graphene and graphene oxide. Chem. Res. Toxicol. 2014, 27, 159–168. [Google Scholar] [CrossRef]
- Bianco, A. Graphene: Safe or toxic? The two faces of the medal. Angew. Chem. Int. Ed. Engl. 2013, 52, 4986–4997. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Yang, S.-T.; Liu, J.-H.; Dong, E.; Wang, Y.; Cao, A.; Liu, Y.; Wang, H. In vitro toxicity evaluation of graphene oxide on A549 cells. Toxicol. Lett. 2011, 200, 201–210. [Google Scholar] [PubMed]
- Sanchez, V.C.; Jachak, A.; Hurt, R.H.; Kane, A.B. Biological Interactions of graphene-family nanomaterials: An interdisciplinary review. Chem. Res. Toxicol. 2012, 25, 15–34. [Google Scholar] [PubMed]
- Hu, W.; Peng, C.; Lv, M.; Li, X.; Zhang, Y.; Chen, N.; Fan, C.; Huang, Q. Protein corona-mediated mitigation of cytotoxicity of graphene oxide. ACS Nano 2011, 5, 3693–3700. [Google Scholar] [CrossRef] [PubMed]
- Cohen-Tanugi, D.; Grossman, J.C. Water desalination across nanoporous graphene. Nano Lett. 2012, 12, 602–608. [Google Scholar] [CrossRef] [PubMed]
- Kottakis, F.; Polytarchou, C.; Foltopoulou, P.; Sanidas, I.; Kampranis, S.C.; Tsichlis, P.N. FGF-2 regulates cell proliferation, migration and angiogenesis through an NDY1/KDM2B-miR-101-EZH2 pathway. Mol. Cell 2011, 43, 285–298. [Google Scholar] [CrossRef] [PubMed]
- Murakami, M.; Nguyen, L.T.; Hatanaka, K.; Schachterle, W.; Chen, P.-Y.; Zhuang, Z.W.; Black, B.L.; Simons, M. FGF-dependent regulation of VEGF receptor 2 expression in mice. J. Clin. Investig. 2011, 121, 2668–2678. [Google Scholar] [CrossRef] [PubMed]
- Delage, B.; Fennell, D.A.; Nicholson, L.; McNeish, I.; Lemoine, N.R.; Crook, T.; Schlosarek, P.W. Arginine deprivation and argininosuccinate synthetase expression in the treatment of cancer. Int. J. Cancer 2010, 126, 2762–2772. [Google Scholar] [CrossRef] [PubMed]
- Wierzbicki, M.; Sawosz, E.; Grodzik, M.; Prasek, M.; Jaworski, S.; Chwalibog, A. Comparison of anti-angiogenic properties of pristine carbon nanoparticles. Nanoscale Res. Lett. 2013, 8, 195. [Google Scholar] [CrossRef] [PubMed]
- Bae, D.-G.; Gho, Y.-S.; Yoon, W.-H.; Chae, C.-B. Arginine-rich anti-vascular endothelial growth factor peptides inhibit tumor growth and metastasis by blocking angiogenesis. J. Biol. Chem. 2000, 275, 13588–13596. [Google Scholar] [CrossRef] [PubMed]
- Yeh, C.-L.; Pai, M.-H.; Li, C.-C.; Tsai, Y.-L.; Yeh, S.-L. Effect of arginine on angiogenesis induced by human colon cancer: In vitro and in vivo studies. J. Nutr. Biochem. 2010, 21, 538–543. [Google Scholar] [CrossRef] [PubMed]
- Kettenmann, H.; Hanisch, U.K.; Verkhratsky, A. Physiology of microglia. Physiol. Rev. 2011, 91, 461–553. [Google Scholar] [CrossRef] [PubMed]
- Ross, D.; Kepa, J.K.; Winski, S.; Beall, H.D.; Anwar, A.; Siegel, D. NAD(P)H:Quinone oxidoreductase 1 (NQO1): Chemoprotection, bioactivation, gene regulation and genetic polymorphisms. Chem. Biol. Interact. 2000, 129, 77–97. [Google Scholar] [CrossRef]
- Qiu, Y.; Wang, Z.; Owens, A.C.; Kulaots, I.; Chen, Y.; Kane, A.B.; Hurt, R.H. Antioxidant chemistry of graphene-based materials and its role in oxidation protection technology. Nanoscale 2014, 6, 11744–11755. [Google Scholar] [CrossRef] [PubMed]
- Dinkova-Kostova, A.T.; Talay, P. NAD(P)H:Quinone acceptor oxidoreductase 1 (NQO1), a multifunctional antioxidant enzyme and exceptionally versatile cytoprotector. Arch. Biochem. Biophys. 2010, 501, 116–123. [Google Scholar] [CrossRef] [PubMed]
- Kung, C.P.; Khaku, S.; Jennis, M.; Murphy, M.E. Identification of TRIML2, a novel p53 target, that enhances p53 SUMOylation and regulates the transactivation of pro-apoptotic genes. Mol. Cancer Res. 2015, 13, 250–262. [Google Scholar] [CrossRef] [PubMed]
- Bonini, P.; Cicconi, S.; Cardinale, A.; Vitale, C.; Serafino, A.L.; Ciotti, M.T.; Marlier, N.J.-L. Oxidative stress induces p53-mediated apoptosis in glia: p53 transcription- independent way to die. J. Neurosci. Res. 2004, 75, 83–95. [Google Scholar] [CrossRef] [PubMed]
- Meulmeester, E.; Jochemsen, A.G. p53: A guide to apoptosis. Curr. Cancer Drug Targets 2008, 8, 87–97. [Google Scholar] [CrossRef] [PubMed]
- Vassilev, L.T.; Vu, B.T.; Graves, B.; Carvajal, D.; Podlaski, F.; Filipovic, Z.; Kong, N.; Kammlott, U.; Lukacs, C.; Klein, C.; et al. In vivo activation of the p53 pathway by small-molecule antagonists of MDM2. Science 2004, 303, 844–848. [Google Scholar] [CrossRef] [PubMed]
- Ventura, A.; Kirsch, D.G.; McLaughlin, M.E.; Tuveson, D.A.; Grimm, J.; Lintault, L.; Newman, J.; Reczek, E.E.; Weissleder, R.; Jacks, T. Restoration of p53 function leads to tumour regression in vivo. Nature 2007, 8, 661–665. [Google Scholar] [CrossRef] [PubMed]
- Lei, H.Z.; Mi, L.J.; Zhou, X.J.; Lei, H.; Mi, L.; Zhou, X.; Chen, J.; Hu, J.; Guo, S.; Zhang, Y. Adsorption of double-stranded DNA to graphene oxide preventing enzymatic digestion. Nanoscale 2011, 3, 3888–3892. [Google Scholar] [CrossRef] [PubMed]
- Green, D.R.; Reed, J.C. Mitochondria and apoptosis. Science 1998, 281, 1309–1312. [Google Scholar] [CrossRef] [PubMed]
- Haupt, S.; Berger, M.; Goldberg, Z.; Haupt, Y. Apoptosis—The p53 network. J. Cell Sci. 2003, 116, 4077–4085. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawosz, E.; Jaworski, S.; Kutwin, M.; Vadalasetty, K.P.; Grodzik, M.; Wierzbicki, M.; Kurantowicz, N.; Strojny, B.; Hotowy, A.; Lipińska, L.; et al. Graphene Functionalized with Arginine Decreases the Development of Glioblastoma Multiforme Tumor in a Gene-Dependent Manner. Int. J. Mol. Sci. 2015, 16, 25214-25233. https://doi.org/10.3390/ijms161025214
Sawosz E, Jaworski S, Kutwin M, Vadalasetty KP, Grodzik M, Wierzbicki M, Kurantowicz N, Strojny B, Hotowy A, Lipińska L, et al. Graphene Functionalized with Arginine Decreases the Development of Glioblastoma Multiforme Tumor in a Gene-Dependent Manner. International Journal of Molecular Sciences. 2015; 16(10):25214-25233. https://doi.org/10.3390/ijms161025214
Chicago/Turabian StyleSawosz, Ewa, Sławomir Jaworski, Marta Kutwin, Krishna Prasad Vadalasetty, Marta Grodzik, Mateusz Wierzbicki, Natalia Kurantowicz, Barbara Strojny, Anna Hotowy, Ludwika Lipińska, and et al. 2015. "Graphene Functionalized with Arginine Decreases the Development of Glioblastoma Multiforme Tumor in a Gene-Dependent Manner" International Journal of Molecular Sciences 16, no. 10: 25214-25233. https://doi.org/10.3390/ijms161025214
APA StyleSawosz, E., Jaworski, S., Kutwin, M., Vadalasetty, K. P., Grodzik, M., Wierzbicki, M., Kurantowicz, N., Strojny, B., Hotowy, A., Lipińska, L., Jagiełło, J., & Chwalibog, A. (2015). Graphene Functionalized with Arginine Decreases the Development of Glioblastoma Multiforme Tumor in a Gene-Dependent Manner. International Journal of Molecular Sciences, 16(10), 25214-25233. https://doi.org/10.3390/ijms161025214