Transcriptome Profiling of Louisiana iris Root and Identification of Genes Involved in Lead-Stress Response
Abstract
:1. Introduction
2. Results and Discussion
2.1. Sequencing and Reads Assembly
| Items | Number |
|---|---|
| Total unigenes | 313,958 |
| Max contig length (bp) | 15,986 |
| Min contig length (bp) | 205 |
| Whole dataset length (bp) | 156,000,000 |
| Average contig length (bp) | 497 |
| N50 | 572 |

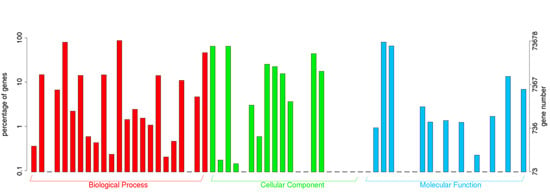
2.2. Annotation of Unigenes
| Annotation Database | Number of Annotations | Percent of Annotation (%) |
|---|---|---|
| Total unigene | 313,958 | - |
| Nr | 89,738 | 28.58% |
| SWISS-PROT | 61,342 | 19.54% |
| TrEMBL | 93,087 | 29.65% |
| CDD | 55,710 | 17.74% |
| PFAM | 80,756 | 25.72% |
| KOG | 43,015 | 13.70% |



| Pathway | Id | Number of DEGs with Pathway Annotations (13,204) | Number of Genes with Pathway Annotations (14,105) | p-Value |
|---|---|---|---|---|
| Metabolism of xenobiotics by cytochrome P450 | ko00980 | 134 (1.02%) | 287 (2.03%) | 3.06 × 10−8 |
| Drug metabolism-cytochrome P450 | ko00982 | 122 (0.92%) | 267 (1.89%) | 4.53 × 10−7 |
| PPAR signaling pathway | ko03320 | 221 (1.67%) | 548 (3.89%) | 9.40 × 10−7 |
| Protein processing in endoplasmic reticulum | ko04141 | 796 (6.03%) | 2332 (16.53%) | 8.32 × 10−6 |
| Ribosome biogenesis in eukaryotes | ko03008 | 264 (2.00%) | 705 (5.00%) | 5.85 × 10−5 |
| RNA polymerase | ko03020 | 122 (0.92%) | 297 (2.11%) | 1.94 × 10−4 |
| Ubiquitin mediated proteolysis | ko04120 | 392 (2.97%) | 1123 (7.06%) | 5.06 × 10−4 |
| Proteasome | ko03050 | 313 (2.37%) | 886 (6.28%) | 9.74 × 10−4 |
| ABC transporters | ko02010 | 152 (1.15%) | 397 (2.81%) | 9.80 × 10−4 |
| Peroxisome | ko04146 | 326 (2.47%) | 929 (6.59%) | 1.11 × 10−3 |
| Glutathione metabolism | ko00480 | 248 (1.88%) | 697 (4.94%) | 2.38 × 10−3 |
| Alanine, aspartate and glutamate metabolism | ko00250 | 210 (1.59) | 594 (4.21%) | 8.02 × 10−3 |
| Pathway | Id | Number of DEGs with Pathway Annotations (13,204) | Number of Genes with Pathway Annotations (14,105) | p-Value |
|---|---|---|---|---|
| Ribosome | ko03010 | 3988 (30.20%) | 8912 (63.18%) | 8.16 × 10−101 |
| Citrate cycle (TCA cycle) | ko00020 | 566 (4.29%) | 1125 (7.98%) | 1.48 × 10−25 |
| Regulation of actin cytoskeleton | ko04810 | 584 (4.42%) | 1262 (8.95%) | 3.45 × 10−16 |
| Gap junction | ko04540 | 269 (2.04%) | 527 (3.74%) | 1.67 × 10−13 |
| Propanoate metabolism | ko00640 | 342 (2.59%) | 722 (5.12%) | 2.32 × 10−11 |
| MAPK signaling pathway | ko04010 | 440 (3.33%) | 1016 (7.20%) | 7.54 × 10−8 |
| Calcium signaling pathway | ko04020 | 284 (2.15%) | 661 (4.69%) | 5.06 × 10−4 |
| d-Glutamine and d-glutamate metabolism | ko00471 | 33 (0.25%) | 58 (0.41%) | 8.42 × 10−4 |
2.3. Validation by Real-Time RT-PCR
| Transcript ID | Description | Fold by RNA-Seq | Fold by qPCR |
|---|---|---|---|
| comp152611_c0_seq1 | Glutathione γ-glutamylcysteinyltransferase | 2.64 | 1.89 |
| comp154333_c1_seq2 | Metallothionein-like protein type 2 | 1.90 | 3.68 |
| comp100849_c0_seq1 | WRKY transcription factor 22 | 0.44 | 0.62 |
| comp123900_c0_seq1 | EREBP-like factor | 1.26 | 1.55 |
| comp145580_c0_seq1 | NAC domain-containing protein 90 | 3.58 | 5.22 |
| comp160851_c0_seq1 | Metal transporter Nramp2 | 1.29 | 1.43 |
| comp142858_c0_seq3 | Nitrate transporter 1.2 | 0.12 | 0.05 |
| comp147199_c0_seq1 | Glutathione S-transferase | 6.73 | 8.33 |
| comp130772_c0_seq3 | Ethylene-responsive transcription factor ERF071 | 2.89 | 2.47 |
| comp1017906_c0_seq1 | Zinc finger CCCH domain-containing protein 40 | 9.86 | 8.73 |
| comp162326_c2_seq1 | Myb proto-oncogene protein. plant | 1.27 | 1.38 |
| comp423843_c0_seq1 | Transcription factor bHLH104 | 2.41 | 3.04 |
2.4. Analysis of Differential Gene Expression
3. Experimental Section
3.1. Sampling
3.2. cDNA Library Preparation and Sequencing
3.3. Transcriptome Sequencing Results Analysis
3.4. Real-Time RT-PCR
3.5. Analysis of Gene Differential Expression
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Sanayei, Y.; Ismail, N.; Talebi, S. Determination of heavy metals in Zayandeh Rood river, Isfahan-Iran. World Appl. Sci. J. 2009, 6, 1209–1214. [Google Scholar]
- Belyaeva, E.A.; Sokolova, T.V.; Emelyanova, L.V.; Zakharova, I.O. Mitochondrial electron transport chain in heavy metal-induced neurotoxicity: Effects of cadmium, mercury, and copper. Sci. World J. 2012, 136063, 14. [Google Scholar] [CrossRef] [PubMed]
- Järup, L. Hazards of heavy metal contamination. Br. Med. Bull. 2003, 68, 167–182. [Google Scholar] [CrossRef] [PubMed]
- Pandey, L.K.; Kumar, D.; Yadav, A.; Rai, J.; Gaur, J. Morphological abnormalities in periphytic diatoms as a tool for biomonitoring of heavy metal pollution in a river. Ecol. Indic. 2014, 36, 272–279. [Google Scholar] [CrossRef]
- Ballerini, E.; Mockaitis, K.; Arnold, M. Transcriptome sequencing and phylogenetic analysis of floral and leaf MIKCC MADS-box and R2R3 MYB transcription factors from the monocot Iris fulva. Gene 2013, 531, 337–346. [Google Scholar] [CrossRef] [PubMed]
- Van Zandt, P.; Mopper, S. Delayed and carryover effects of salinity on flowering in Iris hexagona (Iridaceae). Am. J. Bot. 2002, 89, 1847–1851. [Google Scholar] [CrossRef] [PubMed]
- Nagalakshmi, U.; Wang, Z.; Waern, K.; Shou, C.; Raha, D.; Gerstein, M.; Snyder, M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 2008, 320, 1344–1349. [Google Scholar] [CrossRef] [PubMed]
- Fan, X.; Wang, J.; Yang, N.; Dong, Y.; Liu, L.; Wang, F.; Wang, N.; Chen, H.; Liu, W.; Sun, Y. Gene expression profiling of soybean leaves and roots under salt, saline–alkali and drought stress by high-throughput Illumina sequencing. Gene 2013, 512, 392–402. [Google Scholar] [CrossRef] [PubMed]
- Shen, Y.; Zhang, Y.; Chen, J.; Lin, H.; Zhao, M.; Peng, H.; Liu, L.; Yuan, G.; Zhang, S.; Zhang, Z. Genome expression profile analysis reveals important transcripts in maize roots responding to the stress of heavy metal Pb. Physiol. Plant. 2013, 147, 270–282. [Google Scholar] [CrossRef] [PubMed]
- Wang, G.; Zhu, Q.; Meng, Q.; Wu, C. Transcript profiling during salt stress of young cotton (Gossypium hirsutum) seedlings via Solexa sequencing. Acta Physiol. Plant. 2012, 34, 107–115. [Google Scholar] [CrossRef]
- Wang, X.; Liu, Y.; Jia, Y.; Gu, H.; Ma, H.; Yu, T.; Zhang, H.; Chen, Q.; Ma, L.; Gu, A. Transcriptional responses to drought stress in root and leaf of chickpea seedling. Mol. Biol. Rep. 2012, 39, 8147–8158. [Google Scholar] [CrossRef] [PubMed]
- Yu, S.; Zhang, F.; Yu, Y.; Zhang, D.; Zhao, X.; Wang, W. Transcriptome profiling of dehydration stress in the Chinese cabbage (Brassica rapa L. ssp. pekinensis) by tag sequencing. Plant Mol. Biol. Rep. 2012, 30, 17–28. [Google Scholar] [CrossRef]
- Nachappa, P.; Levy, J.; Tamborindeguy, C. Transcriptome analyses of Bactericera cockerelli adults in response to “Candidatus Liberibacter solanacearum” infection. Mol. Genet. Genom. 2012, 287, 803–817. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Y.; Gao, F.; Liu, R.; Feng, J.; Li, H. De novo sequencing and analysis of root transcriptome using 454 pyrosequencing to discover putative genes associated with drought tolerance in Ammopiptanthus mongolicus. BMC Genom. 2012, 13, 266. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Xu, L.; Chen, Y.; Shen, H.; Gong, Y.; Limera, C.; Liu, L. Transcriptome profiling of radish (Raphanus sativus L.) root and identification of genes involved in response to Lead (Pb) stress with next generation sequencing. PLoS ONE 2013, 8, e66539. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Liu, S.; Guan, H.; Ma, L.; Chen, Z.; Gu, H.; Qu, L. Transcriptional profiling of Arabidopsis seedlings in response to heavy metal lead (Pb). Environ. Exp. Bot. 2009, 67, 377–386. [Google Scholar] [CrossRef]
- Zhou, Y.; Huang, S.; Yu, S.; Gu, J.; Zhao, J.; Han, Y.; Fu, J. The physiological response and sub-cellular localization of lead and cadmium in Iris pseudacorus L. Ecotoxicology 2010, 19, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Huang, S.; Gu, J.; Qiu, S.; Chen, J. Tolerance and accumulation of lead by species of Iris L. Ecotoxicology 2008, 17, 853–859. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Liu, L.; Zhao, Y.; Deng, Y.; Zhu, X.; Huang, S. Overexpression of Iris. lactea var. chinensis metallothionein llMT2a enhances cadmium tolerance in Arabidopsis thaliana. Ecotoxicol. Environ. Saf. 2014, 105, 22–28. [Google Scholar] [PubMed]
- Xu, J.; Sun, J.; Du, L.; Liu, X. Comparative transcriptome analysis of cadmium responses in Solanum nigrum and Solanum torvum. New Phytol. 2012, 196, 110–124. [Google Scholar] [CrossRef] [PubMed]
- Amemiya, Y.; Usami, T.; Okazaki, R.; Katori, M.; Uematsu, S. A new Phytophthora sp. causing basal rot on Japanese Iris. Phytopathology 2011, 101, S6. [Google Scholar]
- Li, J.; Fu, Y.; Pike, S.M.; Bao, J.; Tian, W.; Zhang, Y.; Chen, C.; Zhang, Y.; Li, H.; Huang, J. The Arabidopsis nitrate transporter NRT1.8 functions in nitrate removal from the xylem sap and mediates cadmium tolerance. Plant Cell 2010, 22, 1633–1646. [Google Scholar] [CrossRef] [PubMed]
- Zhu, X.; Tian, S.; Huang, S.; Yuan, H. Effects of Pb on growth and subcellular structure of “Louisiana iris”, and its localization in the plant. Fresenius Environ. Bull. 2014, 23, 2395–2400. [Google Scholar]
- Liu, Z.; Gu, C.; Chen, F.; Yang, D.; Wu, K.; Chen, S.; Jiang, J.; Zhang, Z. Heterologous expression of a Nelumbo nucifera phytochelatin synthase gene enhances cadmium tolerance in Arabidopsis thaliana. Appl. Biochem. Biotechnol. 2012, 166, 722–734. [Google Scholar] [CrossRef] [PubMed]
- Xia, Y.; Qi, Y.; Yuan, Y.; Wang, G.; Cui, J.; Chen, Y.; Zhang, H.; Shen, Z. Overexpression of Elsholtzia haichowensis metallothionein1 (EhMT1) in tobacco plants enhances copper tolerance and accumulation in root cytoplasm and decreases hydrogen peroxide production. J. Hazard. Mater. 2012, 233–234, 65–71. [Google Scholar]
- Gupta, D.; Vandenhove, H.; Inouhe, M. Role of phytochelatins in heavy metal stress and detoxification mechanisms in plants. In Heavy Metal Stress in Plants; Springer: Berlin, Heidelberg, 2013; pp. 73–94. [Google Scholar]
- Yi, Y.; Atagana, H.; Liu, J.; Wu, W.; Wu, S. cDNA sequence encoding metallothionein protein from Aegiceras corniculatum and its gene expression induced by Pb2+ and Cd2+ stresses. Environ. Monit. Assess. 2013, 185, 10201–10208. [Google Scholar]
- An, Z.; Li, C.; Zu, Y.; Du, Y.; Wachter, A.; Gromes, R.; Rausch, T. Expression of BjMT2, a metallothionein 2 from Brassica juncea, increases copper and cadmium tolerance in Escherichia coli and Arabidopsis thaliana, but inhibits root elongation in Arabidopsis thaliana seedlings. J. Exp. Bot. 2006, 57, 3575–3582. [Google Scholar]
- Lv, Y.; Deng, X.; Quan, L.; Xia, Y.; Shen, Z. Metallothioneins BcMT1 and BcMT2 from Brassica campestris enhance tolerance to cadmium and copper and decrease production of reactive oxygen species in Arabidopsis thaliana. Plant Soil 2012, 367, 507–519. [Google Scholar] [CrossRef]
- Xu, J.; Zhu, Y.; Ge, Q.; Li, Y.; Sun, J.; Zhang, Y.; Liu, X. Comparative physiological responses of Solanum nigrum and Solanum torvum to cadmium stress. New Phytol. 2012, 196, 125–138. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.; Naya, L.; Gay, M.; Abian, J.; Becana, M. Functional characterization of an unusual phytochelatin synthase, LjPCS3, of Lotus japonicus. Plant Physiol. 2008, 148, 536–545. [Google Scholar] [CrossRef] [PubMed]
- Ramos, J.; Clemente, M.R.; Naya, L.; Loscos, J.; Perez-Rontome, C.; Sato, S.; Tabata, S.; Becana, M. Phytochelatin synthases of the model legume Lotus japonicus. A small multigene family with differential response to cadmium and alternatively spliced variants. Plant Physiol. 2007, 143, 1110. [Google Scholar] [CrossRef] [PubMed]
- Satoh-Nagasawa, N.; Mori, M.; Nakazawa, N.; Kawamoto, T.; Nagato, Y.; Sakurai, K.; Takahashi, H.; Watanabe, A.; Akagi, H. Mutations in rice (Oryza sativa) heavy metal ATPase 2 (OsHMA2) restrict the translocation of zinc and cadmium. Plant Cell Physiol. 2012, 53, 213–224. [Google Scholar] [CrossRef] [PubMed]
- Kazemi-Dinan, A.; Thomaschky, S.; Stein, R.J.; Krämer, U.; Müller, C. Zinc and cadmium hyperaccumulation act as deterrents towards specialist herbivores and impede the performance of a generalist herbivore. New Phytol. 2014, 202, 628–639. [Google Scholar]
- Moons, A. Ospdr9, which encodes a PDR-type ABC transporter, is induced by heavy metals, hypoxic stress and redox perturbations in rice roots. FEBS Lett. 2003, 553, 370–376. [Google Scholar] [CrossRef]
- Kim, D.Y.; Bovet, L.; Maeshima, M.; Martinoia, E.; Lee, Y. The ABC transporter AtPDR8 is a cadmium extrusion pump conferring heavy metal resistance. Plant J. 2007, 50, 207–218. [Google Scholar] [CrossRef] [PubMed]
- Liu, J.; Ma, J.; He, C.; Li, X.; Zhang, W.; Xu, F.; Lin, Y.; Wang, L. Inhibition of cadmium ion uptake in rice (Oryza sativa) cells by a wall-bound form of silicon. New Phytol. 2013, 200, 691–699. [Google Scholar] [CrossRef] [PubMed]
- Ramesh, S.A.; Shin, R.; Eide, D.J.; Schachtman, D.P. Differential metal selectivity and gene expression of two zinc transporters from rice. Plant Physiol. 2003, 133, 126–134. [Google Scholar] [CrossRef] [PubMed]
- Ishimaru, Y.; Masuda, H.; Suzuki, M.; Bashi, K.; Takahashi, M.; Nakanishi, H.; Mori, S.; Nishizawa, N.K. Overexpression of the OsZIP4 zinc transporter confers disarrangement of zinc distribution in rice plants. J. Exp. Bot. 2007, 58, 2909–2915. [Google Scholar] [CrossRef] [PubMed]
- Korshunova, Y.; Eide, D.; Clark, W.; Guerinot, M.; Pakrasi, H. The IRT1 protein from Arabidopsis thaliana is a metal transporter with a broad substrate range. Plant Mol. Biol. 1999, 40, 37–44. [Google Scholar]
- Sancenón, V.; Puig, S.; Mira, H.; Thiele, D.J.; Peñarrubia, L. Identification of a copper transporter family in Arabidopsis thaliana. Plant Mol. Biol. 2003, 51, 577–587. [Google Scholar] [CrossRef] [PubMed]
- Klaumann, S.; Nickolaus, S.D.; Fürst, S.H.; Starck, S.; Schneider, S.; Ekkehard Neuhaus, H.; Trentmann, O. The tonoplast copper transporter COPT5 acts as an exporter and is required for interorgan allocation of copper in Arabidopsis thaliana. New Phytol. 2011, 192, 393–404. [Google Scholar] [CrossRef] [PubMed]
- Drummond, R.; Tutone, A.; Li, Y.; Gardner, R. A putative magnesium transporter AtMRS2–11 is localized to the plant chloroplast envelope membrane system. Plant Sci. 2006, 170, 78–89. [Google Scholar] [CrossRef]
- Gebert, M.; Meschenmoser, K.; Svidová, S.; Weghuber, J.; Schweyen, R.; Eifler, K.; Lenz, H.; Weyand, K.; Knoop, V. A root-expressed magnesium transporter of the MRS2/MGT gene family in Arabidopsis thaliana allows for growth in low-Mg2+ environments. Plant Cell 2009, 21, 4018–4030. [Google Scholar] [CrossRef] [PubMed]
- Eren, E.; Argüello, J.M. Arabidopsis HMA2, a divalent heavy metal-transporting PIB-type ATPase, is involved in cytoplasmic Zn2+ homeostasis. Plant Physiol. 2004, 136, 3712–3723. [Google Scholar] [CrossRef] [PubMed]
- Ueno, D.; Milner, M.J.; Yamaji, N.; Yokosho, K.; Koyama, E.; Clemencia Zambrano, M.; Kaskie, M.; Ebbs, S.; Kochian, L.; Ma, J. Elevated expression of TcHMA3 plays a key role in the extreme Cd tolerance in a Cd-hyperaccumulating ecotype of Thlaspi caerulescens. Plant J. 2011, 66, 852–862. [Google Scholar] [CrossRef] [PubMed]
- Bernard, C.; Roosens, N.; Czernic, P.; Lebrun, M.; Verbruggen, N. A novel CPx-ATPase from the cadmium hyperaccumulator Thlaspi caerulescens. FEBS Lett. 2004, 569, 140–148. [Google Scholar] [CrossRef] [PubMed]
- Andrés-Colás, N.; Sancenón, V.; Rodríguez-Navarro, S.; Mayo, S.; Thiele, D.; Ecker, J.; Puig, S.; Peñarrubia, L. The Arabidopsis heavy metal P-type ATPase HMA5 interacts with metallochaperones and functions in copper detoxification of roots. Plant J. 2006, 45, 225–236. [Google Scholar] [CrossRef] [PubMed]
- Ganesan, G.; Sankararamasubramanian, H.; Harikrishnan, M.; Ashwin, G.; Parida, A. A MYB transcription factor from the grey mangrove is induced by stress and confers NaCl tolerance in tobacco. J. Exp. Bot. 2012, 63, 4549–4561. [Google Scholar] [CrossRef] [PubMed]
- Yuan, Y.; Juan, Z.; Wang, D.; Ling, H. AtbHLH29 of Arabidopsis thaliana is a functional ortholog of tomato FER involved in controlling iron acquisition in strategy I plants. Cell Res. 2005, 15, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Wu, H.; Chen, C.; Du, J.; Liu, H.; Cui, Y.; Zhang, Y.; He, Y.; Wang, Y.; Chu, C.; Feng, Z. Co-Overexpression FIT with AtbHLH38 or AtbHLH39 in Arabidopsis-enhanced cadmium tolerance via increased cadmium sequestration in roots and improved iron homeostasis of shoots. Plant Physiol. 2012, 158, 790–800. [Google Scholar] [CrossRef] [PubMed]
- Sudo, E.; Itouga, M.; Yoshida-Hatanaka, K.; Ono, Y.; Sakakibara, H. Gene expression and sensitivity in response to copper stress in rice leaves. J. Exp. Bot. 2008, 59, 3465–3474. [Google Scholar] [CrossRef] [PubMed]
- Ban, Q.; Liu, G.; Wang, Y. A DREB gene from Limonium bicolor mediates molecular and physiological responses to copper stress in transgenic tobacco. J. Plant Physiol. 2011, 168, 449–458. [Google Scholar] [CrossRef] [PubMed]
- Cao, Y.; Huang, R.; Cao, Z. Effects of Pb stress on the physiological and biochemical traits of maize. J. Maize Sci. 2004, 13, 61–64. [Google Scholar]
- Grabherr, M.; Haas, B.; Yassour, M.; Levin, J.; Thompson, D.; Amit, I.; Adiconis, X.; Fan, L.; Raychowdhury, R.; Zeng, Q. Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 2011, 29, 644–652. [Google Scholar] [CrossRef] [PubMed]
- Pertea, G.; Huang, X.; Liang, F.; Antonescu, V.; Sultana, R.; Karamycheva, S.; Lee, Y.; White, J.; Cheung, F.; Parvizi, B. TIGR Gene Indices clustering tools (TGICL): A software system for fast clustering of large EST datasets. Bioinformatics 2003, 19, 651–652. [Google Scholar] [CrossRef] [PubMed]
- Gu, C.; Liu, L.; Xu, C.; Zhao, Y.; Zhu, X.; Huang, S. Reference gene selection for quantitative real-time RT-PCR normalization in Iris lactea var. chinensis roots under cadmium, lead, and salt stress conditions. Sci. World J. 2014, 532713, 7. [Google Scholar]
- Gu, C.; Liu, L.; Deng, Y.; Zhu, X.; Lu, X.; Huang, S. Validation of reference genes for RT-qPCR normalization in Iris. lactea var. chinensis leaves under different experimental conditions. Sci. Hortic. 2014, 175, 144–149. [Google Scholar]
- Gu, C.; Zhang, X.; Jiang, J.; Guan, Z.; Zhao, S.; Fang, W.; Liao, Y.; Chen, S.; Chen, F. Chrysanthemum CmNAR2 interacts with CmNRT2 in the control of nitrate uptake. Sci. Rep. 2014, 4, 5833. [Google Scholar] [CrossRef] [PubMed]
- Mortazavi, A.; Williams, B.; McCue, K.; Schaeffer, L.; Wold, B. Mapping and quantifying mammalian transcriptomes by RNA-Seq. Nat. Methods 2008, 5, 621–628. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons by Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Tian, S.; Gu, C.; Liu, L.; Zhu, X.; Zhao, Y.; Huang, S. Transcriptome Profiling of Louisiana iris Root and Identification of Genes Involved in Lead-Stress Response. Int. J. Mol. Sci. 2015, 16, 28087-28097. https://doi.org/10.3390/ijms161226084
Tian S, Gu C, Liu L, Zhu X, Zhao Y, Huang S. Transcriptome Profiling of Louisiana iris Root and Identification of Genes Involved in Lead-Stress Response. International Journal of Molecular Sciences. 2015; 16(12):28087-28097. https://doi.org/10.3390/ijms161226084
Chicago/Turabian StyleTian, Songqing, Chunsun Gu, Liangqin Liu, Xudong Zhu, Yanhai Zhao, and Suzhen Huang. 2015. "Transcriptome Profiling of Louisiana iris Root and Identification of Genes Involved in Lead-Stress Response" International Journal of Molecular Sciences 16, no. 12: 28087-28097. https://doi.org/10.3390/ijms161226084
APA StyleTian, S., Gu, C., Liu, L., Zhu, X., Zhao, Y., & Huang, S. (2015). Transcriptome Profiling of Louisiana iris Root and Identification of Genes Involved in Lead-Stress Response. International Journal of Molecular Sciences, 16(12), 28087-28097. https://doi.org/10.3390/ijms161226084