A Dynamic Combinatorial Approach for Identifying Side Groups that Stabilize DNA-Templated Supramolecular Self-Assemblies
Abstract
:1. Introduction

2. Results and Discussion
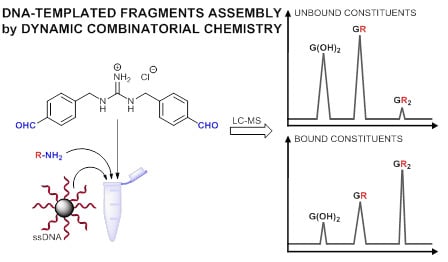
2.1. Design

2.2. Methodology
2.3. Results
| Conditions | MilliQ H2O | Phosphate Buffer | MilliQ H2O | Phosphate Buffer |
|---|---|---|---|---|
| Template | Cellulose | Cellulose | dTn on Cellulose | dTn on Cellulose |
| SA/R (solution) | 3.5 ± 0.4 | 8.1 ± 1.2 | 3.5 ± 0.4 | 8.1 ± 1.2 |
| SA/R (supernatant) | 3.0 ± 0.2 | 10.4 ± 3.3 | 0.09 ± 0.01 | 1.3 |
| SA/R (wash) | 4.2 ± 0.5 | 7.6 ± 1.2 | 2.7 ± 0.5 | 5.1 |
| RW/S | 1.4 ± 0.1 | 0.9 ± 0.2 | 31.4 ± 3.7 | 3.8 |
2.3.1. Parallel Screening of Side Groups



2.3.2. In Situ Screening of Side Groups through Component Selection

3. Experimental Section
3.1. General Procedures and Materials
3.1.1. Materials
3.1.2. HPLC and LC-MS
3.1.3. Mass Spectrometry
3.2. General Procedure for the Generation and Analyses of Dynamic Combinatorial Libraries
3.2.1. Reductive Amination Reactions in Solution
3.2.2. Reductive Amination Reactions with Solid Support
3.2.3. Reductive Amination Reactions with Two Different Amines in Competition
3.2.4. Isolation of Bis-Aminated Product Gua-Benz2
3.2.5. Isolation of Bis-Reduced Product Gua(OH)2 and Bis-Aminated Product Gua-Agmatine2
4. Conclusions
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Schneider, H.J.; Yatsimirsky, A.K. Selectivity in supramolecular host-guest complexes. Chem. Soc. Rev. 2008, 37, 263–277. [Google Scholar] [CrossRef] [PubMed]
- Schneider, H.J. Ligand binding to nucleic acids and proteins: Does selectivity increase with strength? Eur. J. Med. Chem. 2008, 43, 2307–2315. [Google Scholar] [CrossRef] [PubMed]
- Granzhan, A.; Kotera, N.; Teulade-Fichou, M.P. Finding needles in a basestack: Recognition of mismatched base pairs in DNA by small molecules. Chem. Soc. Rev. 2014, 43, 3630–3665. [Google Scholar] [CrossRef] [PubMed]
- Granzhan, A.; Largy, E.; Saettel, N.; Teulade-Fichou, M.P. Macrocyclic DNA-mismatch-binding ligands: Structural determinants of selectivity. Chem. Eur. J. 2010, 16, 878–889. [Google Scholar] [CrossRef] [PubMed]
- Bahr, M.; Gabelica, V.; Granzhan, A.; Teulade-Fichou, M.P.; Weinhold, E. Selective recognition of pyrimidine–pyrimidine DNA mismatches by distance-constrained macrocyclic bis-intercalators. Nucleic Acids Res. 2008, 36, 5000–5012. [Google Scholar] [CrossRef] [PubMed]
- Fechter, E.J.; Olenyuk, B.; Dervan, P.B. Design of a sequence-specific DNA bisintercalator. Angew. Chem. Int. Ed. Engl. 2004, 43, 3591–3594. [Google Scholar] [CrossRef] [PubMed]
- Slamaschwok, A.; Teulade-Fichou, M.P.; Vigneron, J.P.; Taillandier, E.; Lehn, J.M. Selective binding of a macrocyclic bisacridine to DNA hairpins. J. Am. Chem. Soc. 1995, 117, 6822–6830. [Google Scholar] [CrossRef]
- Ranjan, N.; Davis, E.; Xue, L.; Arya, D.P. Dual recognition of the human telomeric G-quadruplex by a neomycin–anthraquinone conjugate. Chem. Commun. 2013, 49, 5796–5798. [Google Scholar] [CrossRef]
- Arambula, J.F.; Ramisetty, S.R.; Baranger, A.M.; Zimmerman, S.C. A simple ligand that selectively targets CUG trinucleotide repeats and inhibits MBNL protein binding. Proc. Natl. Acad. Sci. USA 2009, 106, 16068–16073. [Google Scholar] [CrossRef] [PubMed]
- Madsen, C.S.; Witzke, S.; Kumar, P.; Negi, K.; Sharma, P.K.; Petersen, M.; Nielsen, P. Additional base-pair formation in DNA duplexes by a double-headed nucleotide. Chem. Eur. J. 2012, 18, 7434–7442. [Google Scholar] [CrossRef] [PubMed]
- Haug, R.; Kramer, M.; Richert, C. Three-pronged probes: High-affinity DNA binding with cap, β-alanines and oligopyrrolamides. Chem. Eur. J. 2013, 19, 15822–15826. [Google Scholar] [CrossRef] [PubMed]
- Marlin, F.; Simon, P.; Saison-Behmoaras, T.; Gioyannangeli, C. Delivery of oligonucleotides and analogues: The oligonucleotide conjugate-based approach. Chembiochem 2010, 11, 1493–1500. [Google Scholar] [CrossRef] [PubMed]
- Noir, R.; Kotera, M.; Pons, B.; Remy, J.S.; Behr, J.P. Oligonucleotide–oligospermine conjugates (zip nucleic acids): A convenient means of finely tuning hybridization temperatures. J. Am. Chem. Soc. 2008, 130, 13500–13505. [Google Scholar] [CrossRef] [PubMed]
- Robles, J.; Rajur, S.B.; McLaughlin, L.W. A parallel-stranded DNA triplex tethering a Hoechst 33258 analogue results in complex stabilization by simultaneous major groove and minor groove binding. J. Am. Chem. Soc. 1996, 118, 5820–5821. [Google Scholar] [CrossRef]
- Sinyakov, A.N.; Lokhov, S.G.; Kutyavin, I.V.; Gamper, H.B.; Meyer, R.B. Exceptional and selective stabilization of A–T rich DNA–DNA duplexes by N-methylpyrrole carboxamide peptides conjugated to oligodeoxynucleotides. J. Am. Chem. Soc. 1995, 117, 4995–4996. [Google Scholar] [CrossRef]
- Thuong, N.T.; Asseline, U.; Roig, V.; Takasugi, M.; Helene, C. Oligo(α-deoxynucleotide)s covalently linked to intercalating agents-differential binding to ribopolynucleotides and deoxyribopolynucleotides and stability towards nuclease digestion. Proc. Natl. Acad. Sci. USA 1987, 84, 5129–5133. [Google Scholar] [CrossRef] [PubMed]
- Muse, O.; Zengeya, T.; Mwaura, J.; Hnedzko, D.; McGee, D.W.; Grewer, C.T.; Rozners, E. Sequence selective recognition of double-stranded rna at physiologically relevant conditions using PNA-peptide conjugates. ACS Chem. Biol. 2013, 8, 1683–1686. [Google Scholar] [CrossRef] [PubMed]
- Fechter, E.J.; Dervan, P.B. Allosteric inhibition of protein-DNA complexes by polyamide-intercalator conjugates. J. Am. Chem. Soc. 2003, 125, 8476–8485. [Google Scholar] [CrossRef] [PubMed]
- Szewczyk, J.W.; Baird, E.E.; Dervan, P.B. Sequence-specific recognition of DNA by a major and minor groove binding ligand. Angew. Chem. Int. Ed. Engl. 1996, 35, 1487–1489. [Google Scholar] [CrossRef]
- Tran, T.; Childs-Disney, J.L.; Liu, B.; Guan, L.R.; Rzuczek, S.; Disney, M.D. Targeting the r(CGG) repeats that cause fxtas with modularly assembled small molecules and oligonucleotides. ACS Chem. Biol. 2014, 9, 904–912. [Google Scholar] [CrossRef] [PubMed]
- Childs-Disney, J.L.; Parkesh, R.; Nakamori, M.; Thornton, C.A.; Disney, M.D. Rational design of bioactive, modularly assembled aminoglycosides targeting the RNA that causes myotonic dystrophy type 1. ACS Chem. Biol. 2012, 7, 1984–1993. [Google Scholar] [CrossRef] [PubMed]
- Childs-Disney, J.L.; Tsitovich, P.B.; Disney, M.D. Using modularly assembled ligands to bind RNA internal loops separated by different distances. Chembiochem 2011, 12, 2143–2146. [Google Scholar] [CrossRef] [PubMed]
- Lee, M.M.; Pushechnikov, A.; Disney, M.D. Rational and modular design of potent ligands targeting the RNA that causes myotonic dystrophy 2. ACS Chem. Biol. 2009, 4, 345–355. [Google Scholar] [CrossRef] [PubMed]
- El-Sagheer, A.H.; Brown, T. Combined nucleobase and backbone modifications enhance DNA duplex stability and preserve biocompatibility. Chem. Sci. 2014, 5, 253–259. [Google Scholar] [CrossRef]
- Wojciechowski, F.; Hudson, R.H.E. Fluorescence and hybridization properties of peptide nucleic acid containing a substituted phenylpyrrolocytosine designed to engage guanine with an additional H-bond. J. Am. Chem. Soc. 2008, 130, 12574–12575. [Google Scholar] [CrossRef] [PubMed]
- Quinn, J.R.; Zimmerman, S.C. Structure–function studies on a synthetic guanosine receptor that simultaneously binds Watson–Crick and hoogsteen sites. J. Org. Chem. 2005, 70, 7459–7467. [Google Scholar] [CrossRef] [PubMed]
- Wilds, C.J.; Maier, M.A.; Tereshko, V.; Manoharan, M.; Egli, M. Direct observation of a cytosine analogue that forms five hydrogen bonds to guanosine: Guanidino G-clamp. Angew. Chem. Int. Ed. Engl. 2002, 41, 115–117. [Google Scholar] [CrossRef] [PubMed]
- Lin, K.Y.; Matteucci, M.D. A cytosine analogue capable of clamp-like binding to a guanine in helical nucleic acids. J. Am. Chem. Soc. 1998, 120, 8531–8532. [Google Scholar] [CrossRef]
- Flanagan, W.M.; Wolf, J.J.; Olson, P.; Grant, D.; Lin, K.Y.; Wagner, R.W.; Matteucci, M.D. A cytosine analog that confers enhanced potency to antisense oligonucleotides. Proc. Natl. Acad. Sci. USA 1999, 96, 3513–3518. [Google Scholar] [CrossRef] [PubMed]
- Lou, C.; Dallmann, A.; Marafini, P.; Gao, R.; Brown, T. Enhanced H-bonding and π-stacking in DNA: A potent duplex-stabilizing and mismatch sensing nucleobase analogue. Chem. Sci. 2014, 5, 3836–3844. [Google Scholar] [CrossRef]
- Joly, J.P.; Mata, G.; Eldin, P.; Briant, L.; Fontaine-Vive, F.; Duca, M.; Benhida, R. Artificial nucleobase-amino acid conjugates: A new class of tar RNA binding agents. Chem. Eur. J. 2014, 20, 2071–2079. [Google Scholar] [CrossRef] [PubMed]
- Duca, M.; Malnuit, V.; Barbault, F.; Benhida, R. Design of novel RNA ligands that bind stem-bulge HIV-1 TAR RNA. Chem. Commun. 2010, 46, 6162–6164. [Google Scholar] [CrossRef]
- Rowan, S.J.; Cantrill, S.J.; Cousins, G.R.L.; Sanders, J.K.M.; Stoddart, J.F. Dynamic covalent chemistry. Angew. Chem. Int. Ed. Engl. 2002, 41, 898–952. [Google Scholar] [CrossRef] [PubMed]
- Ulrich, S.; Dumy, P. Probing secondary interactions in biomolecular recognition by dynamic combinatorial chemistry. Chem. Commun. 2014, 50, 5810–5825. [Google Scholar] [CrossRef]
- Herrmann, A. Dynamic combinatorial/covalent chemistry: A tool to read, generate and modulate the bioactivity of compounds and compound mixtures. Chem. Soc. Rev. 2014, 43, 1899–1933. [Google Scholar] [CrossRef] [PubMed]
- Barboiu, M. (Ed.) Topics in Current Chemistry. In Constitutional Dynamic Chemistry; Springer Berlin Heidelberg: Berlin, Germany, 2012; Volume 322.
- Cougnon, F.B.L.; Sanders, J.K.M. Evolution of dynamic combinatorial chemistry. Acc. Chem. Res. 2012, 45, 2211–2221. [Google Scholar] [CrossRef] [PubMed]
- Li, J.W.; Nowak, P.; Otto, S. Dynamic combinatorial libraries: From exploring molecular recognition to systems chemistry. J. Am. Chem. Soc. 2013, 135, 9222–9239. [Google Scholar] [CrossRef] [PubMed]
- Miller, B.L. (Ed.) Dynamic Combinatorial Chemistry in Drug Discovery, Bioorganic Chemistry, and Materials Science; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2010.
- Reek, J.N.H.; Otto, S. (Eds.) Dynamic Combinatorial Chemistry; Wiley-VCH: Weinheim, Germany, 2010.
- Ladame, S. Dynamic combinatorial chemistry: On the road to fulfilling the promise. Org. Biomol. Chem. 2008, 6, 219–226. [Google Scholar] [CrossRef] [PubMed]
- Corbett, P.T.; Leclaire, J.; Vial, L.; West, K.R.; Wietor, J.L.; Sanders, J.K.M.; Otto, S. Dynamic combinatorial chemistry. Chem. Rev. 2006, 106, 3652–3711. [Google Scholar] [CrossRef] [PubMed]
- Lehn, J.M. Dynamic combinatorial chemistry and virtual combinatorial libraries. Chem. Eur. J. 1999, 5, 2455–2463. [Google Scholar] [CrossRef]
- Karan, C.; Miller, B.L. RNA-selective coordination complexes identified via dynamic combinatorial chemistry. J. Am. Chem. Soc. 2001, 123, 7455–7456. [Google Scholar] [CrossRef] [PubMed]
- Klekota, B.; Hammond, M.H.; Miller, B.L. Generation of novel DNA-binding compounds by selection and amplification from self-assembled combinatorial libraries. Tetrahedron Lett. 1997, 38, 8639–8642. [Google Scholar] [CrossRef]
- Gareiss, P.C.; Sobczak, K.; McNaughton, B.R.; Palde, P.B.; Thornton, C.A.; Miller, B.L. Dynamic combinatorial selection of molecules capable of inhibiting the (CUG) repeat RNA-MBNL1 interaction in vitro: Discovery of lead compounds targeting myotonic dystrophy (DM1). J. Am. Chem. Soc. 2008, 130, 16254–16261. [Google Scholar] [CrossRef] [PubMed]
- Whitney, A.M.; Ladame, S.; Balasubramanian, S. Templated ligand assembly by using G-quadruplex DNA and dynamic covalent chemistry. Angew. Chem. Int. Ed. Engl. 2004, 43, 1143–1146. [Google Scholar] [CrossRef] [PubMed]
- Ladame, S.; Whitney, A.M.; Balasubramanian, S. Targeting nucleic acid secondary structures with polyamides using an optimized dynamic combinatorial approach. Angew. Chem. Int. Ed. Engl. 2005, 44, 5736–5739. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Senin, P.; Gomez-Pinto, I.; Grandas, A.; Marchan, V. Identification of ligands for the Tau exon 10 splicing regulatory element rna by using dynamic combinatorial chemistry. Chem. Eur. J. 2011, 17, 1946–1953. [Google Scholar] [CrossRef] [PubMed]
- Aldaye, F.A.; Palmer, A.L.; Sleiman, H.F. Assembling materials with DNA as the guide. Science 2008, 321, 1795–1799. [Google Scholar] [CrossRef] [PubMed]
- Sargsyan, G.; Leonard, B.M.; Kubelka, J.; Balaz, M. Supramolecular ssDNA templated porphyrin and metalloporphyrin nanoassemblies with tunable helicity. Chem. Eur. J. 2014, 20, 1878–1892. [Google Scholar] [CrossRef] [PubMed]
- Sezi, S.; Wagenknecht, H.A. DNA-templated formation of fluorescent self-assembly of ethynyl pyrenes. Chem. Commun. 2013, 49, 9257–9259. [Google Scholar] [CrossRef]
- Lin, J.B.; Surin, M.; Beljonne, D.; Lou, X.W.; van Dongen, J.L.J.; Schenning, A.P.H.J. On the mechanism of dynamic polymerization via recycled ss-DNA templated assembly of non-natural bases. Chem. Sci. 2012, 3, 2732–2736. [Google Scholar] [CrossRef]
- McLaughlin, C.K.; Hamblin, G.D.; Sleiman, H.F. Supramolecular DNA assembly. Chem. Soc. Rev. 2011, 40, 5647–5656. [Google Scholar] [CrossRef] [PubMed]
- Ruiz-Carretero, A.; Janssen, P.G.A.; Kaeser, A.; Schenning, A.P.H.J. DNA-templated assembly of dyes and extended π-conjugated systems. Chem. Commun. 2011, 47, 4340–4347. [Google Scholar] [CrossRef]
- Surin, M.; Janssen, P.G.A.; Lazzaroni, R.; Leclere, P.; Meijer, E.W.; Schenning, A.P.H.J. Supramolecular organization of ssDNA-templated π-conjugated oligomers via hydrogen bonding. Adv. Mater. 2009, 21, 1126–1130. [Google Scholar] [CrossRef]
- Janssen, P.G.A.; Jabbari-Farouji, S.; Surin, M.; Vila, X.; Gielen, J.C.; de Greef, T.F.A.; Vos, M.R.J.; Bomans, P.H.H.; Sommerdijk, N.A.J.M.; Christianen, P.C.M.; et al. Insights into templated supramolecular polymerization: Binding of naphthalene derivatives to ssDNA templates of different lengths. J. Am. Chem. Soc. 2009, 131, 1222–1231. [Google Scholar] [CrossRef] [PubMed]
- Janssen, P.G.A.; Vandenbergh, J.; van Dongen, J.L.J.; Meijer, E.W.; Schenning, A.P.H.J. ssDNA templated self-assembly of chromophores. J. Am. Chem. Soc. 2007, 129, 6078–6079. [Google Scholar] [CrossRef] [PubMed]
- Iwaura, R.; Hoeben, F.J.M.; Masuda, M.; Schenning, A.P.H.J.; Meijer, E.W.; Shimizu, T. Molecular-level helical stack of a nucleotide-appended oligo(p-phenylenevinylene) directed by supramolecular self-assembly with a complementary oligonucleotide as a template. J. Am. Chem. Soc. 2006, 128, 13298–13304. [Google Scholar] [CrossRef] [PubMed]
- Su, W.; Bonnard, V.; Burley, G.A. DNA-templated photonic arrays and assemblies: Design principles and future opportunities. Chem. Eur. J. 2011, 17, 7982–7991. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Eckert, M.A.; Ali, M.M.; Liu, L.; Kang, D.K.; Chang, E.; Pone, E.J.; Sender, L.S.; Fruman, D.A.; Zhao, W. DNA-scaffolded multivalent ligands to modulate cell function. Chembiochem 2014, 15, 1268–1273. [Google Scholar] [CrossRef] [PubMed]
- Sadhu, K.K.; Rothlingshofer, M.; Winssinger, N. DNA as a platform to program assemblies with emerging functions in chemical biology. Israel J. Chem. 2013, 53, 75–86. [Google Scholar] [CrossRef]
- Englund, E.A.; Wang, D.Y.; Fujigaki, H.; Sakai, H.; Micklitsch, C.M.; Ghirlando, R.; Martin-Manso, G.; Pendrak, M.L.; Roberts, D.D.; Durell, S.R.; et al. Programmable multivalent display of receptor ligands using peptide nucleic acid nanoscaffolds. Nat. Commun. 2012, 3, 614. [Google Scholar] [CrossRef] [PubMed]
- Liu, R.; Jiang, B.; Yu, H.Y.; Chaput, J.C. Generating DNA synbodies from previously discovered peptides. ChemBioChem 2011, 12, 1813–1817. [Google Scholar] [CrossRef] [PubMed]
- Paolantoni, D.; Rubio-Magnieto, J.; Cantel, S.; Martinez, J.; Dumy, P.; Surin, M.; Ulrich, S. Probing the importance of π-stacking interactions in DNA-templated self-assembly of bisfunctionalized guanidinium compounds. Chem. Commun. 2014, 50, 14257–14260. [Google Scholar] [CrossRef]
- Gorska, K.; Winssinger, N. Reactions templated by nucleic acids: More ways to translate oligonucleotide-based instructions into emerging function. Angew. Chem. Int. Ed. Engl. 2013, 52, 6820–6843. [Google Scholar] [CrossRef] [PubMed]
- Percivalle, C.; Bartolo, J.F.; Ladame, S. Oligonucleotide-templated chemical reactions: Pushing the boundaries of a nature-inspired process. Org. Biomol. Chem. 2013, 11, 16–26. [Google Scholar]
- Li, X.; Liu, D.R. DNA-templated organic synthesis: Nature’s strategy for controlling chemical reactivity applied to synthetic molecules. Angew. Chem. Int. Ed. Engl. 2004, 43, 4848–4870. [Google Scholar]
- Kleiner, R.E.; Brudno, Y.; Birnbaum, M.E.; Liu, D.R. DNA-templated polymerization of side-chain-functionalized peptide nucleic acid aldehydes. J. Am. Chem. Soc. 2008, 130, 4646–4659. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.Y.; Zhang, Z.Y.J.; Knipe, R.; Lynn, D.G. DNA-catalyzed polymerization. J. Am. Chem. Soc. 2002, 124, 746–747. [Google Scholar] [CrossRef] [PubMed]
- Bouillon, C.; Paolantoni, D.; Rote, J.C.; Bessin, Y.; Peterson, L.W.; Dumy, P.; Ulrich, S. Degradable hybrid materials based on cationic acylhydrazone dynamic covalent polymers promote DNA complexation through multivalent interactions. Chem. Eur. J. 2014, 20, 14705–14714. [Google Scholar] [CrossRef] [PubMed]
- Borch, R.F.; Bernstei., M.D.; Durst, H.D. Cyanohydridoborate anion as a selective reducing agent. J. Am. Chem. Soc. 1971, 93, 2897–2904. [Google Scholar] [CrossRef]
- Benedetti, E.; Duchemin, N.; Bethge, L.; Vonhoff, S.; Klussmann, S.; Vasseur, J.J.; Cossy, J.; Smietana, M.; Arseniyadis, S. DNA-cellulose: An economical, fully recyclable and highly effective chiral biomaterial for asymmetric catalysis. Chem. Commun. 2015. [Google Scholar] [CrossRef]
- Misuraca, M.C.; Moulin, E.; Ruff, Y.; Giuseppone, N. Experimental and theoretical methods for the analyses of dynamic combinatorial libraries. New J. Chem. 2014, 38, 3336–3349. [Google Scholar] [CrossRef]
- Besenius, P.; Cormack, P.A.G.; Ludlow, R.F.; Otto, S.; Sherrington, D.C. Affinity chromatography in dynamic combinatorial libraries: One-pot amplification and isolation of a strongly binding receptor. Org. Biomol. Chem. 2010, 8, 2414–2418. [Google Scholar] [CrossRef] [Green Version]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Paolantoni, D.; Cantel, S.; Dumy, P.; Ulrich, S. A Dynamic Combinatorial Approach for Identifying Side Groups that Stabilize DNA-Templated Supramolecular Self-Assemblies. Int. J. Mol. Sci. 2015, 16, 3609-3625. https://doi.org/10.3390/ijms16023609
Paolantoni D, Cantel S, Dumy P, Ulrich S. A Dynamic Combinatorial Approach for Identifying Side Groups that Stabilize DNA-Templated Supramolecular Self-Assemblies. International Journal of Molecular Sciences. 2015; 16(2):3609-3625. https://doi.org/10.3390/ijms16023609
Chicago/Turabian StylePaolantoni, Delphine, Sonia Cantel, Pascal Dumy, and Sébastien Ulrich. 2015. "A Dynamic Combinatorial Approach for Identifying Side Groups that Stabilize DNA-Templated Supramolecular Self-Assemblies" International Journal of Molecular Sciences 16, no. 2: 3609-3625. https://doi.org/10.3390/ijms16023609
APA StylePaolantoni, D., Cantel, S., Dumy, P., & Ulrich, S. (2015). A Dynamic Combinatorial Approach for Identifying Side Groups that Stabilize DNA-Templated Supramolecular Self-Assemblies. International Journal of Molecular Sciences, 16(2), 3609-3625. https://doi.org/10.3390/ijms16023609