Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria
Abstract
:1. Introduction

2. Results and Discussion
2.1. Structures of Adult Ascaris suum QFR and Porcine Succinate-Ubiquinone Reductase (SQR)


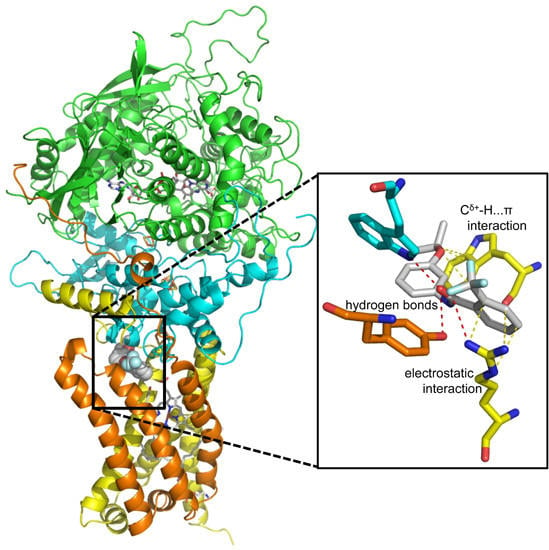
2.2. Structures of Flutolanil Binding Sites of A. suum QFR and Porcine SQR


2.2.1. Interaction between Trifluoromethylbenzene Ring and Arginine Residue
| Derivative | R1 | R2 | IC50 (QFR/SQR, μM) | PDB Code (QFR/SQR) |
|---|---|---|---|---|
| flutolanil | –CF3 | –O–CH(CH3)2 | 0.0581/45.9 | 5C2T/4YXD |
| 1 | –I | –O–CH(CH3)2 | 0.0723/6.43 | 4YSZ/3AE7 |
| 2 | –CH3 | –O–CH(CH3)2 | 0.515/90.0 | 4YT0/– |
| 3 | –CF3 | –CH2–N(CH3)2 | 3.42/256 | –/3AEA |
| 4 | –CF3 | –O–C6H5 | 0.0794/16.2 | –/– |
| 5 | –CF3 | –C6H5 | 0.0245/8.61 | 4YTM/3ABV |
| 6 | –CF3 | –O–C6H5F5 | 0.330/236 | –/3AE9 |
2.2.2. C–H…π Interaction between Isopropoxybenzene Ring and Tryptophan Residue
2.3. Structures of A. suum QFR and Porcine SQR in Complexes with Flutolanil Derivatives

2.4. Inhibitors with Higher Potency and Specificity

3. Experimental Section
3.1. Purification
3.2. Crystallization
3.3. X-ray Diffraction Data Collection, Structure Determination and Refinement
3.4. Enzyme Assays
4. Conclusions
Supplementary Information


| Data Set | QFR-RQ | QFR-Flutolanil | QFR-Derivative 1 | QFR-Derivative 2 | QFR-Derivative 5 | QFR-NN23 | QFR-NN28 |
|---|---|---|---|---|---|---|---|
| Space Group | |||||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 |
| Data Collection | |||||||
| a/b/c (Å) | 122.8, 123.6, 219.8 | 124.3, 131.5, 222.5 | 123.9, 127.0, 219.3 | 122.8, 123.4, 219.0 | 123.7, 126.4, 220.9 | 124.2, 127.9, 220.5 | 124.1, 127.7, 220.7 |
| X-ray source | SPring8 BL44XU | KEK-PF-AR NW12 | KEK-PF-AR NW12 | KEK-PF BL17A | SPring8 BL44XU | SPring8 BL44XU | SPring8 BL44XU |
| Wavelength (Å) | 0.9000 | 1.000 | 1.0000 | 0.98000 | 0.9000 | 0.9000 | 0.9000 |
| Temperature (K) | 100 | 100 | 100 | 100 | 100 | 100 | 100 |
| Resolution (Å) | 50.0–2.75 (2.85–2.75) a | 50–2.9 (2.95–2.9) a | 50.0–3.3 (3.36–3.3) a | 50–3.65 (3.71–3.65) a | 50.00–3.4 (3.52–3.4) a | 50–2.25 (2.29–2.25) a | 50–3.1 (3.15–3.1) a |
| Total reflections | 499,461 | 328,842 | 283,779 | 196,929 | 261,990 | 907,709 | 326,789 |
| Unique reflections | 87,281 | 73,077 | 52,914 | 37,582 | 49,409 | 166,618 | 64,793 |
| Completeness (%) | 99.9 (100.0) a | 95.4 (87.2) a | 98.2 (93.7) a | 97.3 (94.1) a | 95.6 (93.0) a | 96.4 (91.6) a | 96.0 (95.7) a |
| Rmerge I b | 0.092 (0.759) a | 0.096 (0.624) a | 0.137 (0.449) a | 0.164 (0.575) a | 0.136 (0.390) a | 0.082 (0.566) a | 0.114 (0.561) a |
| I/σ(I) | 7.4 (2.2) a | 12.4 (1.3) a | 5.9 (3.3) a | 4.5 (2.4) a | 11.1 (2.7) a | 11.4 (2.9) a | 16.6 (3.0) a |
| Redundancy | 5.7 (5.4) a | 4.5 (3.0) a | 5.5 (4.7) a | 5.4 (4.6) a | 5.5 (3.1) a | 5.7 (4.8) a | 5.3 (4.7) a |
| Refinement | |||||||
| Resolution (Å) | 20–2.75 | 40–2.91 | 30–3.3 | 20–3.66 | 20–3.4 | 20–2.25 | 20–3.1 |
| No. of reflections | 78,102 | 56,377 | 48,822 | 31,772 | 43,572 | 152,442 | 58,730 |
| Rwork c/Rfree d | 0.196/0.252 | 0.204/0.272 | 0.179/0.250 | 0.193/0.262 | 0.178/0.239 | 0.181/0.221 | 0.183/0.238 |
| Number of Atoms | |||||||
| Protein | 17,889 | 17,862 | 17,978 | 17,956 | 17,967 | 18,007 | 17,978 |
| FAD | 106 | 106 | 106 | 106 | 106 | 106 | 106 |
| Malonate (Fumalate) | 14 | 16 | 14 | 16 | 14 | 14 | 14 |
| FeS clusters | 38 | 38 | 38 | 38 | 38 | 38 | 38 |
| Heme | 86 | 86 | 86 | 86 | 86 | 86 | 86 |
| Inhibitor | 44 | 46 | 40 | 40 | 50 | 48 | 44 |
| Lipid | 88 | 88 | 88 | 88 | 44 | 88 | 88 |
| Solvent molecules | 106 | - | 15 | - | - | 721 | 27 |
| B-factors (Å2) | |||||||
| Protein | 50.8 | 86.8 | 62.3 | 59.6 | 79.2 | 49.5 | 66.7 |
| FAD | 28.3 | 61.8 | 42.7 | 31.4 | 50.7 | 35.7 | 49.8 |
| Malonate (Fumalate) | 38.3 | 106.0 | 51.8 | 47.1 | 75.6 | 42.3 | 63.1 |
| FeS clusters | 31.8 | 59.5 | 45.8 | 33.2 | 49.1 | 35.8 | 51.2 |
| Heme | 56.5 | 75.7 | 70.1 | 73.8 | 89.5 | 47.7 | 70.6 |
| Inhibitor | 58.2 | 71.7 | 65.4 | 83.7 | 94.6 | 45.5 | 75.9 |
| Lipid | 71.1 | 119.7 | 110.4 | - | 100.6 | 72.4 | 91.7 |
| Solvent molecules | 30.0 | - | 17.3 | - | - | 44.6 | 43.5 |
| R.m.s. Deviation | |||||||
| Bond length (Å) | 0.008 | 0.013 | 0.008 | 0.006 | 0.007 | 0.009 | 0.007 |
| Bond angle (°) | 1.35 | 1.60 | 1.41 | 1.14 | 1.27 | 1.45 | 1.23 |
| PDB code | 5C2T | 3VRB | 4YSZ | 4YT0 | 4YTM | 4YSX | 4YSY |
| Data Set | SQR-Flutolanil | SQR-Derivative 1 | SQR-Derivative 3 | SQR-Derivative 5 | SQR-Derivative 6 | SQR-NN23 | |||
|---|---|---|---|---|---|---|---|---|---|
| Space Group | |||||||||
| Space group | P212121 | P212121 | P212121 | P212121 | P212121 | P212121 | |||
| Data Collection | |||||||||
| a/b/c (Å) | 70.4, 83.7, 292.6 | 71.2, 84.0, 293.7 | 71.5, 83.8, 295.0 | 71.7, 84.2, 294.4 | 72.6, 84.2, 295.6 | 71.4, 84.0, 294.8 | |||
| X-ray source | SPring8 BL44XU | KEK-PF BL17A | SPring8 BL41XU | KEK-PF-AR NW12 | SPring8 BL41XU | KEK-PF BL17A | |||
| Wavelength (Å) | 0.9000 | 0.98000 | 1.0000 | 1.000 | 1.00000 | 0.9800 | |||
| Temperature (K) | 100 | 100 | 100 | 100 | 100 | 100 | |||
| Resolution (Å) | 50–3.0 (3.05–3.0) a | 50–3.6 (3.66–3.6) a | 50–3.39 (3.51–3.39) | 50–3.2 (3.31–3.2) a | 50–3.3 (3.42–3.3) a | 50–3.1 (3.15–3.1) a | |||
| Total reflections | 184,860 | 123,966 | 172,365 | 139,252 | 109,564 | 142,584 | |||
| Unique reflections | 35,891 | 20,323 | 25,146 | 29,008 | 27,349 | 31,105 | |||
| Completeness (%) | 97.5 (95.1) a | 97.0 (77.8) a | 98.3 (93.9) | 99.8 (100.0) a | 98.2 (99.4) a | 93.3 (94.9) a | |||
| Rmerge I b | 0.099 (0.464) a | 0.149 (0.553) a | 0.091 (0.594) | 0.076 (0.277) a | 0.08 (0.46) a | 0.095 (0.516) a | |||
| I/σ(I) | 11.5 (2.8) a | 16.1 (2.2) a | 8.0 (3.2) | 19.3 (5.2) a | 12.8 (2.0) a | 11.4 (1.7) a | |||
| Redundancy | 5.3 (4.4) a | 6.1(4.4) a | 3.7 (3.7) | 4.8 (4.9) a | 4.0 (4.1) a | 3.9 (4.6) a | |||
| Refinement | |||||||||
| Resolution (Å) | 20–3.0 | 50–3.62 | 50–3.39 | 40–3.24 | 40–3.31 | 20–3.1 | |||
| No. of reflections | 33,058 | 20,118 | 25,081 | 28,950 | 27,275 | 25,461 | |||
| Rwork c/Rfree d | 0.206/0.258 | 0.256/0.305 | 0.236/0.286 | 0.203/0.253 | 0.257/0.307 | 0.208/0.266 | |||
| Number of Atoms | |||||||||
| Protein | 8480 | 8480 | 8480 | 8480 | 8480 | 8480 | |||
| FAD | 53 | 53 | 53 | 53 | 53 | 53 | |||
| Malonate (Fumalate) | – | – | 7 | 7 | 7 | – | |||
| FeS clusters | 19 | 19 | 19 | 19 | 19 | 19 | |||
| Heme | 43 | 43 | 43 | 43 | 43 | 43 | |||
| Inhibitor | 23 | 23 | 23 | 25 | 31 | 24 | |||
| Lipid | – | – | 44 | 44 | 44 | – | |||
| Solvent molecules | – | – | – | – | – | 22 | |||
| B-factors (Å2) | |||||||||
| Protein | 80.6 | 145.3 | 113.9 | 71.0 | 124.7 | 71.1 | |||
| FAD | 70.9 | 127.5 | 84.5 | 56.1 | 100.3 | 58.4 | |||
| Malonate (Fumalate) | – | – | 177.5 | 93.1 | 222.2 | – | |||
| FeS clusters | 64.9 | 124.7 | 84.4 | 55.1 | 94.0 | 54.7 | |||
| Heme | 75.1 | 135.0 | 95.7 | 52.5 | 102.4 | 59.9 | |||
| Inhibitor | 84.1 | 138.0 | 104.1 | 61.7 | 137.5 | 62.7 | |||
| Lipid | – | – | 147.8 | 88.9 | 153.4 | – | |||
| Solvent molecules | – | – | – | – | – | 44.1 | |||
| R.m.s. Deviation | |||||||||
| Bond length (Å) | 0.008 | 0.005 | 0.005 | 0.007 | 0.005 | 0.007 | |||
| Bond angle (°) | 1.31 | 0.84 | 0.88 | 1.08 | 0.867 | 1.24 | |||
| PDB code | 4YXD | 3AE7 | 3AEA | 3ABV | 3AE9 | 4YTP | |||
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Kita, K.; Takamiya, S. Electron-transfer complexes in Ascaris mitochondria. Adv. Parasitol. 2002, 51, 95–131. [Google Scholar] [PubMed]
- Kita, K.; Nihei, C.; Tomitsuka, E. Parasite mitochondria as a drug target: Diversity and dynamic changes during the life cycle. Curr. Med. Chem. 2003, 10, 1241–1253. [Google Scholar] [CrossRef]
- Kita, K.; Shiomi, K.; Õmura, S. Parasitology in Japan: Advances in drug discovery and biochemical studies. Trends Parasitol. 2007, 23, 223–229. [Google Scholar] [CrossRef] [PubMed]
- Kroger, A.; Geisler, V.; Lemma, E.; Theis, F.; Lenger, R. Bacterial fumarate respiration. Arch. Microbiol. 1992, 158, 311–314. [Google Scholar] [CrossRef]
- Saruta, F.; Kuramochi, T.; Nakamura, K.; Takamiya, S.; Yu, Y.; Aoki, T.; Sekimizu, K.; Kojima, S.; Kita, K. Stage-specific isoforms of ComplexII (succinate-ubiquinone oxidoreductase) in mitochondria from the parasitic nematode, Ascaris suum. J. Biol. Chem. 1995, 270, 928–932. [Google Scholar] [CrossRef] [PubMed]
- Kohler, P.; Bachmann, R. Mechanisms of respiration and phosphorylation in Ascaris muscle mitochondria. Mol. Biochem. Parasitol. 1980, 1, 75–90. [Google Scholar] [CrossRef]
- Oya, H.; Kita, K. Comparative Biochemistry of Parasitic Helminths; Bennet, E., Behm, C., Bryant, C., Eds.; Chapman and hall: London, UK, 1998; pp. 35–53. [Google Scholar]
- Tomitsuka, E.; Kita, K.; Esumi, H. Regulation of succinate-ubiquinone reductase and fumarate reductase activities in human complex II by phosphorylation of its flavoprotein subunit. Proc. Jpn. Acad. Ser. B 2009, 85, 258–265. [Google Scholar] [CrossRef]
- Tomitsuka, E.; Kita, K.; Esumi, H. The NADH-fumarate reductase system, a novel mitochondrial energy metabolism, is a new target for anticancer therapy in tumor microenvironments. Ann. N. Y. Acad. Sci. 2010, 1201, 44–49. [Google Scholar] [CrossRef] [PubMed]
- Tomitsuka, E.; Kita, K.; Esumi, H. An anticancer agent, pyrvinium pamoate inhibits the NADH-fumarate reductase system—A unique mitochondrial metabolism in tumor microenvironments. J. Biochem. 2012, 152, 171–183. [Google Scholar] [CrossRef] [PubMed]
- Sakai, C.; Tomitsuka, E.; Miyagishi, M.; Harada, S.; Kita, K. Type II Fp of human mitochondrial respiratory complex II and its role in adaptation to hypoxia and nutrition-deprived conditions. Mitochondrion 2013, 13, 602–609. [Google Scholar] [CrossRef] [PubMed]
- Matsumoto, J.; Sakamoto, K.; Shinjyo, N.; Kido, Y.; Yamamoto, N.; Yagi, K.; Miyoshi, H.; Nonaka, N.; Katakura, K.; Kita, K.; et al. Anaerobic NADH-fumarate reductase system is predominant in the respiratory chain of Echinococcus multilocularis, providing a novel target for the chemotherapy of alveolar echinococcosis. Antimicrob. Agents Chemother. 2008, 52, 164–170. [Google Scholar] [CrossRef] [PubMed]
- Omura, S.; Miyadera, H.; Ui, H.; Shiomi, K.; Yamaguchi, Y.; Masuma, R.; Nagamitsu, T.; Takano, D.; Sunazuka, T.; Harder, A.; et al. An anthelmintic compound, nafuredin, shows selective inhibition of complex I in helminth mitochondria. Proc. Natl. Acad. Sci. USA 2001, 98, 60–62. [Google Scholar] [CrossRef] [PubMed]
- Sakai, C.; Tomitsuka, E.; Esumi, H.; Harada, S.; Kita, K. Mitochondrial fumarate reductase as a target of chemotherapy: From parasites to cancer cells. Biochim. Biophys. Acta-Gen. Subj. 2012, 1820, 643–651. [Google Scholar] [CrossRef] [PubMed]
- Armson, A.; Grubb, W.B.; Mendis, A.H. The effect of electron transport (ET) inhibitors and thiabendazole on the fumarate reductase (FR) and succinate dehydrogenase (SDH) of strongyloides ratti infective (L3) larvae. Int. J. Parasitol. 1995, 25, 261–263. [Google Scholar] [CrossRef]
- Kohler, P.; Bachmann, R. The effects of the antiparasitic drugs levamisole, thiabendazole, praziquantel, and chloroquine on mitochondrial electron transport in muscle tissue from Ascaris suum. Mol. Pharmacol. 1978, 14, 155–163. [Google Scholar] [PubMed]
- Komuniecki, R.; Harris, B.G.; Marr, J.; Mueller, M. Biochemistry and Molecular Biology of Parasites; Academic: London, UK, 1995; pp. 49–66. [Google Scholar]
- Tielens, A.G.; van Hellemond, J.J. The electron transport chain in anaerobically functioning eukaryotes. Biochim. Biophys. Acta 1998, 1365, 71–78. [Google Scholar] [CrossRef]
- Paranagama, M.P.; Sakamoto, K.; Amino, H.; Awano, M.; Miyoshi, H.; Kita, K. Contribution of the FAD and quinone binding sites to the production of reactive oxygen species (ROS) from Ascaris suum mitochondrial complex II. Mitochondrion 2010, 10, 158–165. [Google Scholar] [CrossRef] [PubMed]
- Omura, S.; Tomoda, H.; Kimura, K.; Zhen, D.Z.; Kumagai, H.; Igarashi, K.; Imamura, N.; Takahashi, Y.; Tanaka, Y.; Iwai, Y. Atpenins, new antifungal antibiotics produced by Penicillium sp. Production, isolation, physico-chemical and biological properties. J. Antibiot. Tokyo 1988, 41, 1769–1773. [Google Scholar] [CrossRef] [PubMed]
- Miyadera, H.; Shiomi, K.; Ui, H.; Yamaguchi, Y.; Masuma, R.; Tomoda, H.; Miyoshi, H.; Kita, K.; Osanai, A.; Omura, S. Atpenins, potent and specific inhibitors of mitochondrial complex II (succinate-ubiquinone oxidoreductase). Proc. Natl. Acad. Sci. USA 2003, 100, 473–477. [Google Scholar] [CrossRef] [PubMed]
- Wojtovich, A.P.; Brookes, P.S. The complex II inhibitor atpenin A5 protects against cardiac ischemia-reperfusion injury via activation of mitochondrial KATP channels. Basic Res. Cardiol. 2009, 104, 121–129. [Google Scholar] [CrossRef] [PubMed]
- Osanai, A.; Harada, S.; Sakamoto, K.; Shimizu, H.; Inaoka, D.K.; Kita, K. Crystallization of mitochondrial rhodoquinol-fumarate reductase from the parasitic nematode Ascaris suum with the specific inhibitor flutolanil. Acta Crystallogr. Sect. F 2009, 65, 941–944. [Google Scholar] [CrossRef] [PubMed]
- Yankovskaya, V.; Horsefield, R.; Tornroth, S.; Luna-Chavez, C.; Miyoshi, H.; Leger, C.; Byrne, B.; Cecchini, G.; Iwata, S. Architecture of succinate dehydrogenase and reactive oxygen species generation. Science 2003, 299, 700–704. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.S.; Sun, G.; Cobessi, D.; Wang, A.C.; Shen, J.T.; Tung, E.Y.; Anderson, V.E.; Berry, E.A. 3-Nitropropionic acid is a suicide inhibitor of mitochondrial respiration that, upon oxidation by complex II, forms a covalent adduct with a catalytic base arginine in the active site of the enzyme. J. Biol. Chem. 2006, 281, 5965–5972. [Google Scholar] [CrossRef] [PubMed]
- Sun, F.; Huo, X.; Zhai, Y.; Wang, A.; Xu, J.; Su, D.; Bartlam, M.; Rao, Z. Crystal structure of mitochondrial respiratory membrane protein complex II. Cell 2005, 121, 1043–1057. [Google Scholar] [CrossRef] [PubMed]
- Iverson, T.M.; Luna-Chavez, C.; Cecchini, G.; Rees, D.C. Structure of the Escherichia coli fumarate reductase respiratory complex. Science 1999, 284, 1961–1966. [Google Scholar] [CrossRef] [PubMed]
- Iverson, T.M.; Luna-Chavez, C.; Croal, L.R.; Cecchini, G.; Rees, D.C. Crystallographic studies of the Escherichia coli quinol-fumarate reductase with inhibitors bound to the quinol-binding site. J. Biol. Chem. 2002, 277, 16124–16130. [Google Scholar] [CrossRef] [PubMed]
- Shimizu, H.; Osanai, A.; Sakamoto, K.; Inaoka, D.K.; Shiba, T.; Harada, S.; Kita, K. Crystal structure of mitochondrial quinol-fumarate reductase from the parasitic nematode Ascaris suum. J. Biochem. 2012, 151, 589–592. [Google Scholar] [CrossRef] [PubMed]
- Kuramochi, T.; Hirawake, H.; Kojima, S.; Takamiya, S.; Furushima, R.; Aoki, T.; Komuniecki, R.; Kita, K. Sequence comparison between the flavoprotein subunit of the fumarate reductase (complex II) of the anaerobic parasitic nematode, Ascaris suum and the succinate dehydrogenase of theaerobic, free-living nematode, Caenorhabditis elegans. Mol. Biochem. Parasitol. 1994, 68, 177–187. [Google Scholar] [CrossRef]
- Saruta, F.; Hirawake, H.; Takamiya, S.; Ma, Y.C.; Aoki, T.; Sekimizu, K.; Kojima, S.; Kita, K. Cloning of a cDNA encoding the small subunit of cytochrome b558 (cybS) of mitochondrial fumarate reductase (complex II) from adult Ascaris suum. Biochim. Biophys. Acta 1996, 1276, 1–5. [Google Scholar] [CrossRef]
- Amino, H.; Wang, H.; Hirawake, H.; Saruta, F.; Mizuchi, D.; Mineki, R.; Shindo, N.; Murayama, K.; Takamiya, S.; Aoki, T.; et al. Stage-specific isoforms of Ascaris suum complex II: The fumarate reductase of the parasitic adult and the succinate dehydrogenase of free-living larvae share a common iron-sulfur subunit. Mol. Biochem. Parasitol. 2000, 106, 63–76. [Google Scholar] [CrossRef]
- Iwata, F.; Shinjyo, N.; Amino, H.; Sakamoto, K.; Islam, M.K.; Tsuji, N.; Kita, K. Change of subunit composition of mitochondrial complex II (Succinate-ubiquinone reductase/Quinol-fumarate reductase) in Ascaris suum during the migration in the experimental host. Parasitol. Int. 2008, 57, 54–61. [Google Scholar] [CrossRef] [PubMed]
- Motoba, K.; Uchida, M.; Tada, E. Mode of antifungal action and selectivity of flutolanil. Agric. Biol. Chem. 1988, 52, 1445–1449. [Google Scholar] [CrossRef]
- Ito, Y.; Muraguchi, H.; Seshime, Y.; Oita, S.; Yanagi, S.O. Flutolanil and carboxin resistance in Coprinus cinereus conferred by a mutation in the cytochrome b560 subunit of succinate dehydrogenase complex (Complex II). Mol. Genet. Genomics 2004, 272, 328–335. [Google Scholar] [CrossRef] [PubMed]
- Ernster, L.; Nordenbrand, K. Skeletal muscle mitochondria. Methods Enzymol. 1967, 10, 84–94. [Google Scholar]
- Huo, X.; Su, D.; Wang, A.; Zhai, Y.; Xu, J.; Li, X.; Bartlam, M.; Sun, F.; Rao, Z. Preliminary molecular characterization and crystallization of mitochondrial respiratory complex II from porcine heart. FEBS J. 2007, 274, 1524–1529. [Google Scholar] [CrossRef] [PubMed]
- Otwinowski, Z.; Minor, W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997, 276, 307–326. [Google Scholar]
- Vagin, A.; Teplyakov, A. Molrep: An automated program for molecular replacement. J. Appl. Crystallogr. 1997, 30, 1022–1025. [Google Scholar] [CrossRef]
- Collaborative Computational Project, Number 4. The CCP4 suite: Programs for protein crystallography. Acta Crystallogr. Sect. D 1994, 50, 760–763.
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Inaoka, D.K.; Shiba, T.; Sato, D.; Balogun, E.O.; Sasaki, T.; Nagahama, M.; Oda, M.; Matsuoka, S.; Ohmori, J.; Honma, T.; et al. Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria. Int. J. Mol. Sci. 2015, 16, 15287-15308. https://doi.org/10.3390/ijms160715287
Inaoka DK, Shiba T, Sato D, Balogun EO, Sasaki T, Nagahama M, Oda M, Matsuoka S, Ohmori J, Honma T, et al. Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria. International Journal of Molecular Sciences. 2015; 16(7):15287-15308. https://doi.org/10.3390/ijms160715287
Chicago/Turabian StyleInaoka, Daniel Ken, Tomoo Shiba, Dan Sato, Emmanuel Oluwadare Balogun, Tsuyoshi Sasaki, Madoka Nagahama, Masatsugu Oda, Shigeru Matsuoka, Junko Ohmori, Teruki Honma, and et al. 2015. "Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria" International Journal of Molecular Sciences 16, no. 7: 15287-15308. https://doi.org/10.3390/ijms160715287
APA StyleInaoka, D. K., Shiba, T., Sato, D., Balogun, E. O., Sasaki, T., Nagahama, M., Oda, M., Matsuoka, S., Ohmori, J., Honma, T., Inoue, M., Kita, K., & Harada, S. (2015). Structural Insights into the Molecular Design of Flutolanil Derivatives Targeted for Fumarate Respiration of Parasite Mitochondria. International Journal of Molecular Sciences, 16(7), 15287-15308. https://doi.org/10.3390/ijms160715287