Thermostable Carbonic Anhydrases in Biotechnological Applications
Abstract
:1. Introduction
2. Thermostable CAs in CO2 Capture and Storage Processes


3. Carbonic Anhydrases Isolated from Thermophilic Bacteria
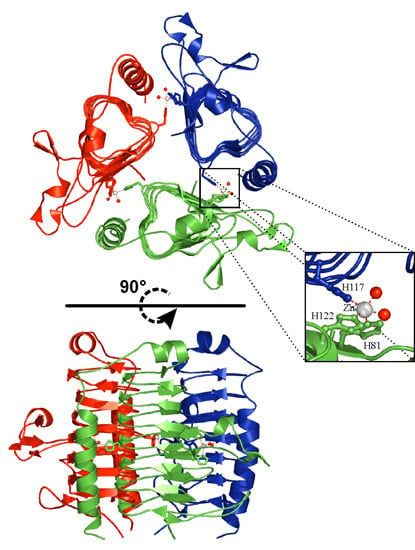
3.1. α-Carbonic Anhydrases
| Enzyme | Class | kcat (s−1) | kcat/KM (M−1·s−1) | Ref. |
|---|---|---|---|---|
| hCA II | α | 1.40 × 106 | 1.5 × 108 | [63] |
| SspCA | α | 9.35 × 105 | 1.1 × 108 | [57] |
| SazCA | α | 4.40 × 106 | 3.5 × 108 | [61] |
| TaCA | α | 1.60 × 106 | 1.6 × 108 | [62] |
| PmCA | α | 3.2 × 105 | 3.0 × 107 | [46] |
| Cab | β | 1.7 × 104 | 5.9 × 106 | [64] |
| MtCam (expressed in E. coli and purified aerobically) | γ | 6.8 × 104 | 3.1 × 106 | [65] |
| MtCam (expressed in E. coli and purified anaerobically) | γ | 24.3 × 104 | 5.4 × 106 | [65] |
| MtCam (expressed in M. acetivorans and purified anaerobically) | γ | 23.1 × 104 | 3.9 × 106 | [66] |
| Temperature (°C) | Half-Life (Days) | |||
|---|---|---|---|---|
| bCA II | SspCA | TaCA | PmCA | |
| 40 | 6 | 53 | 152 | 75 |
| 60 | n.d. | n.d. | 77 | 29 |
| 70 | <1 | 8 | n.d. | n.d. |



3.2. β-Carbonic Anhydrases


3.3. γ-Carbonic Anhydrases



4. Thermostable Carbonic Anhydrases Obtained by Protein Engineering Techniques
5. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Figueroa, J.D.; Fout, T.; Plasynski, S.; McIlvried, H.; Srivastava, R.D. Advances in CO2 capture technology—The U.S. department of energy’s carbon sequestration program. Int. J. Greenh. Gas Control 2008, 2, 9–20. [Google Scholar] [CrossRef]
- Rayalu, S.; Yadav, R.; Wanjari, S.; Prabhu, C.; Mushnoori, S.C.; Labhsetwar, N.; Satyanarayanan, T.; Kotwal, S.; Wate, S.R.; Hong, S.-G.; et al. Nanobiocatalysts for carbon capture, sequestration and valorisation. Top. Catal. 2012, 55, 1217–1230. [Google Scholar] [CrossRef]
- Shekh, A.Y.; Krishnamurthi, K.; Mudliar, S.N.; Yadav, R.R.; Fulke, A.B.; Devi, S.S.; Chakrabarti, T. Recent advancements in carbonic anhydrase-driven processes for CO2 sequestration: Minireview. Crit. Rev. Environ. Sci. Technol. 2012, 42, 1419–1440. [Google Scholar] [CrossRef]
- Benson, S.M.; Surles, T. Carbon dioxide capture and storage: An overview with emphasis on capture and storage in deep geological formations. Proc. IEEE 2006, 94, 1795–1805. [Google Scholar] [CrossRef]
- Zevenhoven, R.; Eloneva, S.; Teir, S. Chemical fixation of CO2 in carbonates: Routes to valuable products and long-term storage. Catal. Today 2006, 115, 73–79. [Google Scholar] [CrossRef]
- Arakawa, H.; Aresta, M.; Armor, J.N.; Barteau, M.A.; Beckman, E.J.; Bell, A.T.; Bercaw, J.E.; Creutz, C.; Dinjus, E.; Dixon, D.A.; et al. Catalysis research of relevance to carbon management: Progress, challenges, and opportunities. Chem. Rev. 2001, 101, 953–996. [Google Scholar] [CrossRef] [PubMed]
- Sakakura, T.; Choi, J.C.; Yasuda, H. Transformation of carbon dioxide. Chem. Rev. 2007, 107, 2365–2387. [Google Scholar] [CrossRef] [PubMed]
- Beckman, E.J. Making polymers from carbon dioxide. Science 1999, 283, 946–947. [Google Scholar] [CrossRef]
- Yang, H.; Xu, Z.; Fan, M.; Gupta, R.; Slimane, R.B.; Bland, A.E.; Wright, I. Progress in carbon dioxide separation and capture: A review. J. Environ. Sci. 2008, 20, 14–27. [Google Scholar] [CrossRef]
- Alterio, V.; di Fiore, A.; D’Ambrosio, K.; Supuran, C.T.; de Simone, G. Multiple binding modes of inhibitors to carbonic anhydrases: How to design specific drugs targeting 15 different isoforms? Chem. Rev. 2012, 112, 4421–4468. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T.; de Simone, G. Carbonic Anhydrases as Biocatalysts-From Theory to Medical and Industrial Applications; Elsevier B.V.: Berlin, Germany, 2015. [Google Scholar]
- De Simone, G.; di Fiore, A.; Capasso, C.; Supuran, C.T. The zinc coordination pattern in the eta-carbonic anhydrase from Plasmodium falciparum is different from all other carbonic anhydrase genetic families. Bioorg. Med. Chem. Lett. 2015, 25, 1385–1389. [Google Scholar] [CrossRef] [PubMed]
- Silverman, D.N.; McKenna, R. Solvent-mediated proton transfer in catalysis by carbonic anhydrase. Acc. Chem. Res. 2007, 40, 669–675. [Google Scholar] [CrossRef] [PubMed]
- Mikulski, R.L.; Silverman, D.N. Proton transfer in catalysis and the role of proton shuttles in carbonic anhydrase. Biochim. Biophys. Acta 2010, 1804, 422–426. [Google Scholar] [CrossRef] [PubMed]
- Boone, C.D.; Pinard, M.; McKenna, R.; Silverman, D. Catalytic mechanism of alpha-class carbonic anhydrases: CO2 hydration and proton transfer. Subcell. Biochem. 2014, 75, 31–52. [Google Scholar] [PubMed]
- Tu, C.K.; Silverman, D.N.; Forsman, C.; Jonsson, B.H.; Lindskog, S. Role of histidine 64 in the catalytic mechanism of human carbonic anhydrase II studied with a site-specific mutant. Biochemistry 1989, 28, 7913–7918. [Google Scholar] [CrossRef] [PubMed]
- Borchert, M.; Saunders, P. Heat Stable Carbonic Anhydrases and Their Use. Patent WO 2008095057 A2, 3 January 2008. [Google Scholar]
- Forsman, C.; Behravan, G.; Osterman, A.; Jonsson, B.H. Production of active human carbonic anhydrase II in E. coli. Acta Chem. Scand. B 1988, 42, 314–318. [Google Scholar] [CrossRef] [PubMed]
- Silverman, D.N. Carbonic anhydrase: Oxygen-18 exchange catalyzed by an enzyme with rate-contributing proton-transfer steps. Methods Enzymol. 1982, 87, 732–752. [Google Scholar] [PubMed]
- Bond, G.M.; Stringer, J.; Brandvold, D.K.; Simsek, F.A.; Medina, M.G.; Egeland, G. Development of integrated system for biomimetic CO2 sequestration using the enzyme carbonic anhydrase. Energy Fuels 2001, 15, 309–316. [Google Scholar] [CrossRef]
- Fisher, Z.; Boone, C.D.; Biswas, S.M.; Venkatakrishnan, B.; Aggarwal, M.; Tu, C.; Agbandje-McKenna, M.; Silverman, D.; McKenna, R. Kinetic and structural characterization of thermostabilized mutants of human carbonic anhydrase II. Protein Eng. Des. Sel. 2012, 25, 347–355. [Google Scholar] [CrossRef] [PubMed]
- Kanbar, B.; Ozdemir, E. Thermal stability of carbonic anhydrase immobilized within polyurethane foam. Biotechnol. Prog. 2010, 26, 1474–1480. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.; Lawal, A.; Stephenson, P.; Sidders, J.; Ramshaw, C. Post-combustion CO2 capture with chemical absorption: A state of the art review. Chem. Eng. Res. Des. 2011, 89, 1609–1624. [Google Scholar] [CrossRef] [Green Version]
- Yu, C.H.; Huang, C.H.; Tan, C.S. A review of CO2 capture by absorption and adsorption. Aerosol Air Qual. Res. 2012, 12, 745–769. [Google Scholar] [CrossRef]
- Huang, H.P.; Shi, Y.; Li, W.; Chang, S.G. Dual alkali approaches for the capture and separation of CO2. Energy Fuels 2001, 15, 263–268. [Google Scholar] [CrossRef]
- Rochelle, G.T. Amine scrubbing for CO2 capture. Science 2009, 325, 1652–1654. [Google Scholar] [CrossRef] [PubMed]
- Puxty, G.; Rowland, R.; Allport, A.; Yang, Q.; Bown, M.; Burns, R.; Maeder, M.; Attalla, M. Carbon dioxide postcombustion capture: A novel screening study of the carbon dioxide absorption performance of 76 amines. Environ. Sci. Technol. 2009, 43, 6427–6433. [Google Scholar] [CrossRef] [PubMed]
- Danckwerts, P.V. The reaction of CO2 with ethanolamines. Chem. Eng. Sci. 1979, 34, 443–446. [Google Scholar] [CrossRef]
- Bishnoi, S.; Rochelle, G.T. Absorption of carbon dioxide into aqueous piperazine: Reaction kinetics, mass transfer and solubility. Chem. Eng. Sci. 2000, 55, 5531–5543. [Google Scholar] [CrossRef]
- Xiao, J.; Li, C.W.; Li, M.H. Kinetics of absorption of carbon dioxide into aqueous solutions of 2-amino-2-methyl-1-propanol plus monoethanolamine. Chem. Eng. Sci. 2000, 55, 161–175. [Google Scholar] [CrossRef]
- Liao, C.H.; Li, M.H. Kinetics of absorption of carbon dioxide into aqueous solutions of monoethanolamine plus N-methyldiethanolamine. Chem. Eng. Sci. 2002, 57, 4569–4582. [Google Scholar] [CrossRef]
- Carson, J.K.; Marsh, K.N.; Mather, A.E. Enthalpy of solution of carbon dioxide in (water + monoethanolamine, or diethanolamine, or N-methyldiethanolamine) and (water + monoethanolamine + N-methyldiethanolamine) at T = 298.15 K. J. Chem. Thermodyn. 2000, 32, 1285–1296. [Google Scholar] [CrossRef]
- Chowdhury, F.A.; Yamada, H.; Higashii, T.; Goto, K.; Onoda, M. CO2 capture by tertiary amine absorbents: A performance comparison study. Ind. Eng. Chem. Res. 2013, 52, 8323–8331. [Google Scholar] [CrossRef]
- Versteeg, G.F.; van Dijck, L.A.J.; van Swaaij, W.P.M. On the kinetics between CO2 and alkanolamines both in aqueous and non-aqueous solutions. An overview. Chem. Eng. Commun. 1996, 144, 113–158. [Google Scholar] [CrossRef]
- Mirjafari, P.; Asghari, K.; Mahinpey, N. Investigating the application of enzyme carbonic anhydrase for CO2 sequestration purposes. Ind. Eng. Chem. Res. 2007, 46, 921–926. [Google Scholar] [CrossRef]
- Vinoba, M.; Bhagiyalakshmi, M.; Grace, A.N.; Kim, D.H.; Yoon, Y.; Nam, S.C.; Baek, I.H.; Jeong, S.K. Carbonic anhydrase promotes the absorption rate of CO2 in post combustion processes. J. Phys. Chem. B 2013, 117, 5683–5690. [Google Scholar] [CrossRef] [PubMed]
- Rossi, M. A New Heat-Stable Carbonic Anhydrase and Uses Thereof. Patent WO 2013064195 A1, 8 May 2014. [Google Scholar]
- Daigle, R.; Madore, E.; Fradette, S. Techniques for CO2 Capture Using Sulfurihydrogenibium sp. Carbonic Anhydrase. Patent WO 2014066999 A1, 8 May 2014. [Google Scholar]
- Borchert, M.; Saunders, P. Heat-Stable Carbonic Anhydrases and Their Use. Patent US 8945826 B2, 3 February 2015. [Google Scholar]
- Borchert, M.; Saunders, P. Heat Stable Carbonic Anhydrases and Their Use. Patent WO 2010151787, 4 February 2010. [Google Scholar]
- Alvizo, O.; Nguyen, L.J.; Savile, C.K.; Bresson, J.A.; Lakhapatri, S.L.; Solis, E.O.; Fox, R.J.; Broering, J.M.; Benoit, M.R.; Zimmerman, S.A.; et al. Directed evolution of an ultrastable carbonic anhydrase for highly efficient carbon capture from flue gas. Proc. Natl. Acad. Sci. USA 2014, 111, 16436–16441. [Google Scholar] [CrossRef] [PubMed]
- Clark, L.; Zimmerman, S. Engineered Beta-Class Carbonic Anhydrase Polypeptides and Uses Thereof. Patent WO 2011066304 A3, 12 January 2012. [Google Scholar]
- Srivastava, S.; Bharti, R.K.; Verma, P.K.; Thakur, I.S. Cloning and expression of gamma carbonic anhydrase from Serratia sp. ISTD04 for sequestration of carbon dioxide and formation of calcite. Bioresour. Technol. 2015, 188, 209–213. [Google Scholar] [CrossRef] [PubMed]
- Sharma, A.; Bhattacharya, A.; Singh, S. Purification and characterization of an extracellular carbonic anhydrase from Pseudomonas fragi. Process Biochem. 2009, 44, 1293–1297. [Google Scholar] [CrossRef]
- Jo, B.H.; Seo, J.H.; Cha, H.J. Bacterial extremo-alpha-carbonic anhydrases from deep-sea hydrothermal vents as potential biocatalysts for CO2 sequestration. J. Mol. Catal. B Enzym. 2014, 109, 31–39. [Google Scholar] [CrossRef]
- Kanth, B.K.; Jun, S.Y.; Kumari, S.; Pack, S.P. Highly thermostable carbonic anhydrase from Persephonella marina EX-H1: Its expression and characterization for CO2 sequestration applications. Process Biochem. 2014, 49, 2114–2121. [Google Scholar] [CrossRef]
- Borchert, M.S. Heat Stable Persephonella Carbonic Anhydrases and Their Use. Patent WO 2012025577, 1 March 2012. [Google Scholar]
- Hyung Joon, C.; Byung Hoon, J.; Jeong Hyun, S. Carbonic Anhydrase Having Thermal Stability and Carbon Dioxide Collection Using the Same. Patent WO 2015056858, 23 April 2015. [Google Scholar]
- Seung Pil, B. Carbonic Anhydrase Isolated from Persephonella Marina and Use Thereof. Patent KR 2014139787, 22 May 2014. [Google Scholar]
- Lee, S.W.; Park, S.B.; Jeong, S.K.; Lim, K.S.; Lee, S.H.; Trachtenberg, M.C. On carbon dioxide storage based on biomineralization strategies. Micron 2010, 41, 273–282. [Google Scholar] [CrossRef] [PubMed]
- Vinoba, M.; Kim, D.H.; Lim, K.S.; Jeong, S.K.; Lee, S.W.; Alagar, M. Biomimetic sequestration of CO2 and reformation to CaCO3 using bovine carbonic anhydrase immobilized on SBA-15. Energy Fuels 2011, 25, 438–445. [Google Scholar] [CrossRef]
- Wanjari, S.; Prabhu, C.; Yadav, R.; Satyanarayana, T.; Labhsetwar, N.; Rayalu, S. Immobilization of carbonic anhydrase on chitosan beads for enhanced carbonation reaction. Process Biochem. 2011, 46, 1010–1018. [Google Scholar] [CrossRef]
- Ozdemir, E. Biomimetic CO2 sequestration: Immobilization of carbonic anhydrase within polyurethane foam. Energy Fuels 2009, 23, 5725–5730. [Google Scholar] [CrossRef]
- Migliardini, F.; de Luca, V.; Carginale, V.; Rossi, M.; Corbo, P.; Supuran, C.T.; Capasso, C. Biomimetic CO2 capture using a highly thermostable bacterial alpha-carbonic anhydrase immobilized on a polyurethane foam. J. Enzym. Inhib. Med. Chem. 2014, 29, 146–150. [Google Scholar] [CrossRef] [PubMed]
- Salemme, F.R.; Weber, P.C. Engineered Carbonic Anhydrase Proteins for CO2 Scrubbing Applications. US 20140178962 A1, 2014. [Google Scholar]
- Borchert, M.; Saunders, P. Heat Stable Carbonic Anhydrases and Their Use. Patent WO 2008095057 A3, 12 February 2009. [Google Scholar]
- Capasso, C.; de Luca, V.; Carginale, V.; Cannio, R.; Rossi, M. Biochemical properties of a novel and highly thermostable bacterial alpha-carbonic anhydrase from Sulfurihydrogenibium yellowstonense YO3AOP1. J. Enzym. Inhib. Med. Chem. 2012, 27, 892–897. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V.; Vullo, D.; Scozzafava, A.; Carginale, V.; Rossi, M.; Supuran, C.T.; Capasso, C. Anion inhibition studies of an alpha-carbonic anhydrase from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Bioorg. Med. Chem. Lett. 2012, 22, 5630–5634. [Google Scholar] [CrossRef] [PubMed]
- Vullo, D.; de Luca, V.; Scozzafava, A.; Carginale, V.; Rossi, M.; Supuran, C.T.; Capasso, C. Anion inhibition studies of the fastest carbonic anhydrase (CA) known, the extremo-CA from the bacterium Sulfurihydrogenibium azorense. Bioorg. Med. Chem. Lett. 2012, 22, 7142–7145. [Google Scholar] [CrossRef] [PubMed]
- Akdemir, A.; Vullo, D.; de Luca, V.; Scozzafava, A.; Carginale, V.; Rossi, M.; Supuran, C.T.; Capasso, C. The extremo-alpha-carbonic anhydrase (CA) from Sulfurihydrogenibium azorense, the fastest CA known, is highly activated by amino acids and amines. Bioorg. Med. Chem. Lett. 2013, 23, 1087–1090. [Google Scholar] [CrossRef] [PubMed]
- De Luca, V.; Vullo, D.; Scozzafava, A.; Carginale, V.; Rossi, M.; Supuran, C.T.; Capasso, C. An alpha-carbonic anhydrase from the thermophilic bacterium Sulphurihydrogenibium azorense is the fastest enzyme known for the CO2 hydration reaction. Bioorg. Med. Chem. 2013, 21, 1465–1469. [Google Scholar] [CrossRef] [PubMed]
- James, P.; Isupov, M.N.; Sayer, C.; Saneei, V.; Berg, S.; Lioliou, M.; Kotlar, H.K.; Littlechild, J.A. The structure of a tetrameric alpha-carbonic anhydrase from Thermovibrio ammonificans reveals a core formed around intermolecular disulfides that contribute to its thermostability. Acta Crystallogr. D Biol. Crystallogr. 2014, 70, 2607–2618. [Google Scholar] [CrossRef] [PubMed]
- Supuran, C.T. Carbonic anhydrases: Novel therapeutic applications for inhibitors and activators. Nat. Rev. Drug Discov. 2008, 7, 168–181. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.S.; Ferry, J.G. A plant-type (beta-class) carbonic anhydrase in the thermophilic methanoarchaeon Methanobacterium thermoautotrophicum. J. Bacteriol. 1999, 181, 6247–6253. [Google Scholar] [PubMed]
- Tripp, B.C.; Bell, C.B., 3rd; Cruz, F.; Krebs, C.; Ferry, J.G. A role for iron in an ancient carbonic anhydrase. J. Biol. Chem. 2004, 279, 6683–6687. [Google Scholar] [CrossRef] [PubMed]
- Macauley, S.R.; Zimmerman, S.A.; Apolinario, E.E.; Evilia, C.; Hou, Y.M.; Ferry, J.G.; Sowers, K.R. The archetype gamma-class carbonic anhydrase (Cam) contains iron when synthesized in vivo. Biochemistry 2009, 48, 817–819. [Google Scholar] [CrossRef] [PubMed]
- Di Fiore, A.; Capasso, C.; de Luca, V.; Monti, S.M.; Carginale, V.; Supuran, C.T.; Scozzafava, A.; Pedone, C.; Rossi, M.; de Simone, G. X-ray structure of the first “extremo-alpha-carbonic anhydrase”, a dimeric enzyme from the thermophilic bacterium Sulfurihydrogenibium yellowstonense YO3AOP1. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1150–1159. [Google Scholar] [CrossRef] [PubMed]
- Russo, M.E.; Olivieri, G.; Capasso, C.; de Luca, V.; Marzocchella, A.; Salatino, P.; Rossi, M. Kinetic study of a novel thermo-stable alpha-carbonic anhydrase for biomimetic CO2 capture. Enzym. Microb. Technol. 2013, 53, 271–277. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Xue, Y.; Sauer-Eriksson, E.; Chirica, L.; Lindskog, S.; Jonsson, B.H. Crystal structure of carbonic anhydrase from Neisseria gonorrhoeae and its complex with the inhibitor acetazolamide. J. Mol. Biol. 1998, 283, 301–310. [Google Scholar] [CrossRef] [PubMed]
- De Simone, G.; Monti, S.M.; Alterio, V.; Buonanno, M.; de Luca, V.; Rossi, M.; Carginale, V.; Supuran, C.T.; Capasso, C.; di Fiore, A. Crystal structure of the most catalytically effective carbonic anhydrase enzyme known, SazCA from the thermophilic bacterium Sulfurihydrogenibium azorense. Bioorg. Med. Chem. Lett. 2015, 25, 2002–2006. [Google Scholar] [CrossRef] [PubMed]
- Krissinel, E.; Henrick, K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007, 372, 774–797. [Google Scholar] [CrossRef] [PubMed]
- Tahirov, T.H.; Oki, H.; Tsukihara, T.; Ogasahara, K.; Yutani, K.; Ogata, K.; Izu, Y.; Tsunasawa, S.; Kato, I. Crystal structure of methionine aminopeptidase from hyperthermophile, Pyrococcus furiosus. J. Mol. Biol. 1998, 284, 101–124. [Google Scholar] [CrossRef] [PubMed]
- Neish, A.C. Studies on chloroplasts: Their chemical composition and the distribution of certain metabolites between the chloroplasts and the remainder of the leaf. Biochem. J. 1939, 33, 300–308. [Google Scholar] [PubMed]
- Burnell, J.N.; Gibbs, M.J.; Mason, J.G. Spinach chloroplastic carbonic anhydrase: Nucleotide sequence analysis of cDNA. Plant Physiol. 1990, 92, 37–40. [Google Scholar] [CrossRef] [PubMed]
- Rowlett, R.S. Structure and catalytic mechanism of the beta-carbonic anhydrases. Biochim. Biophys. Acta 2010, 1804, 362–373. [Google Scholar] [CrossRef] [PubMed]
- Smith, D.R.; Doucette-Stamm, L.A.; Deloughery, C.; Lee, H.; Dubois, J.; Aldredge, T.; Bashirzadeh, R.; Blakely, D.; Cook, R.; Gilbert, K.; et al. Complete genome sequence of Methanobacterium thermoautotrophicum deltaH: Functional analysis and comparative genomics. J. Bacteriol. 1997, 179, 7135–7155. [Google Scholar] [PubMed]
- Strop, P.; Smith, K.S.; Iverson, T.M.; Ferry, J.G.; Rees, D.C. Crystal structure of the “cab”-type beta class carbonic anhydrase from the archaeon Methanobacterium thermoautotrophicum. J. Biol. Chem. 2001, 276, 10299–10305. [Google Scholar] [CrossRef] [PubMed]
- Mitsuhashi, S.; Mizushima, T.; Yamashita, E.; Yamamoto, M.; Kumasaka, T.; Moriyama, H.; Ueki, T.; Miyachi, S.; Tsukihara, T. X-ray structure of beta-carbonic anhydrase from the red alga, Porphyridium purpureum, reveals a novel catalytic site for CO2 hydration. J. Biol. Chem. 2000, 275, 5521–5526. [Google Scholar] [CrossRef] [PubMed]
- Kimber, M.S.; Pai, E.F. The active site architecture of Pisum sativum beta-carbonic anhydrase is a mirror image of that of alpha-carbonic anhydrases. EMBO J. 2000, 19, 1407–1418. [Google Scholar] [CrossRef] [PubMed]
- Cronk, J.D.; Endrizzi, J.A.; Cronk, M.R.; O’Neill, J.W.; Zhang, K.Y. Crystal structure of E. coli beta-carbonic anhydrase, an enzyme with an unusual pH-dependent activity. Protein Sci. 2001, 10, 911–922. [Google Scholar] [CrossRef] [PubMed]
- Cronk, J.D.; Rowlett, R.S.; Zhang, K.Y.; Tu, C.; Endrizzi, J.A.; Lee, J.; Gareiss, P.C.; Preiss, J.R. Identification of a novel noncatalytic bicarbonate binding site in eubacterial beta-carbonic anhydrase. Biochemistry 2006, 45, 4351–4361. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias, A.S.; Bergfors, T.; Jones, T.A.; Hogbom, M. Structural mechanics of the pH-dependent activity of beta-carbonic anhydrase from Mycobacterium tuberculosis. J. Biol. Chem. 2006, 281, 4993–4999. [Google Scholar] [CrossRef] [PubMed]
- Covarrubias, A.S.; Larsson, A.M.; Högbom, M.; Lindberg, J.; Bergfors, T.; Björkelid, C.; Mowbray, S.L.; Unge, T.; Jones, T.A. Structure and function of carbonic anhydrases from Mycobacterium tuberculosis. J. Biol. Chem. 2005, 280, 18782–18789. [Google Scholar] [CrossRef] [PubMed]
- Sawaya, M.R.; Cannon, G.C.; Heinhorst, S.; Tanaka, S.; Williams, E.B.; Yeates, T.O.; Kerfeld, C.A. The structure of beta-carbonic anhydrase from the carboxysomal shell reveals a distinct subclass with one active site for the price of two. J. Biol. Chem. 2006, 281, 7546–7555. [Google Scholar] [CrossRef] [PubMed]
- Smith, K.S.; Jakubzick, C.; Whittam, T.S.; Ferry, J.G. Carbonic anhydrase is an ancient enzyme widespread in prokaryotes. Proc. Natl. Acad. Sci. USA 1999, 96, 15184–15189. [Google Scholar] [CrossRef] [PubMed]
- Ferry, J.G. Carbonic Anhydrases of environmentally and medically relevant anaerobic prokaryotes. In CARBONIC ANHYDRASES AS BIOCATALYSTS-From Theory to Medical and Industrial Applications; Supran, C.T., de Simone, G., Eds.; Elsevier B.V.: Berilin, Germany, 2015; pp. 325–336. [Google Scholar]
- Ferry, J.G. The gamma class of carbonic anhydrases. Biochim. Biophys. Acta 2010, 1804, 374–381. [Google Scholar] [CrossRef] [PubMed]
- Alber, B.E.; Ferry, J.G. A carbonic anhydrase from the archaeon Methanosarcina thermophila. Proc. Natl. Acad. Sci. USA 1994, 91, 6909–6913. [Google Scholar] [CrossRef] [PubMed]
- Alber, B.E.; Ferry, J.G. Characterization of heterologously produced carbonic anhydrase from Methanosarcina thermophila. J. Bacteriol. 1996, 178, 3270–3274. [Google Scholar] [PubMed]
- Alber, B.E.; Colangelo, C.M.; Dong, J.; Stalhandske, C.M.; Baird, T.T.; Tu, C.; Fierke, C.A.; Silverman, D.N.; Scott, R.A.; Ferry, J.G. Kinetic and spectroscopic characterization of the gamma-carbonic anhydrase from the methanoarchaeon Methanosarcina thermophila. Biochemistry 1999, 38, 13119–13128. [Google Scholar] [CrossRef] [PubMed]
- Kisker, C.; Schindelin, H.; Alber, B.E.; Ferry, J.G.; Rees, D.C. A left-hand beta-helix revealed by the crystal structure of a carbonic anhydrase from the archaeon Methanosarcina thermophila. EMBO J. 1996, 15, 2323–2330. [Google Scholar] [PubMed]
- Iverson, T.M.; Alber, B.E.; Kisker, C.; Ferry, J.G.; Rees, D.C. A closer look at the active site of gamma-class carbonic anhydrases: High-resolution crystallographic studies of the carbonic anhydrase from Methanosarcina thermophila. Biochemistry 2000, 39, 9222–9231. [Google Scholar] [CrossRef] [PubMed]
- Hutchinson, E.G.; Thornton, J.M. PROMOTIF—A program to identify and analyze structural motifs in proteins. Protein Sci. 1996, 5, 212–220. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.A.; Ferry, J.G. Proposal for a hydrogen bond network in the active site of the prototypic gamma-class carbonic anhydrase. Biochemistry 2006, 45, 5149–5157. [Google Scholar] [CrossRef] [PubMed]
- Zimmerman, S.; Domsic, J.F.; Tu, C.; Robbins, A.H.; McKenna, R.; Silverman, D.N.; Ferry, J.G. Role of Trp19 and Tyr200 in catalysis by the gamma-class carbonic anhydrase from Methanosarcina thermophila. Arch. Biochem. Biophys. 2013, 529, 11–17. [Google Scholar] [CrossRef] [PubMed]
- Jeyakanthan, J.; Rangarajan, S.; Mridula, P.; Kanaujia, S.P.; Shiro, Y.; Kuramitsu, S.; Yokoyama, S.; Sekar, K. Observation of a calcium-binding site in the gamma-class carbonic anhydrase from Pyrococcus horikoshii. Acta Crystallogr. D Biol. Crystallogr. 2008, 64, 1012–1019. [Google Scholar] [CrossRef] [PubMed]
- Peña, K.L.; Castel, S.E.; de Araujo, C.; Espie, G.S.; Kimber, M.S. Structural basis of the oxidative activation of the carboxysomal gamma-carbonic anhydrase, CcmM. Proc. Natl. Acad. Sci. USA 2010, 107, 2455–2460. [Google Scholar] [CrossRef] [PubMed]
- Boone, C.D.; Habibzadegan, A.; Tu, C.; Silverman, D.N.; McKenna, R. Structural and catalytic characterization of a thermally stable and acid-stable variant of human carbonic anhydrase II containing an engineered disulfide bond. Acta Crystallogr. D Biol. Crystallogr. 2013, 69, 1414–1422. [Google Scholar] [CrossRef] [PubMed]
- Daigle, R.; Desrochers, M. Carbonic Anhydrase Mutants Having Increased Stability under High Temperature Conditions. Patent US 7521217, 21 April 2009. [Google Scholar]
- Fisher, S.Z.; Tu, C.; Bhatt, D.; Govindasamy, L.; Agbandje-McKenna, M.; McKenna, R.; Silverman, D.N. Speeding up proton transfer in a fast enzyme: Kinetic and crystallographic studies on the effect of hydrophobic amino acid substitutions in the active site of human carbonic anhydrase II. Biochemistry 2007, 46, 3803–3813. [Google Scholar] [CrossRef] [PubMed]
- Mikulski, R.; West, D.; Sippel, K.H.; Avvaru, B.S.; Aggarwal, M.; Tu, C.; McKenna, R.; Silverman, D.N. Water networks in fast proton transfer during catalysis by human carbonic anhydrase II. Biochemistry 2013, 52, 125–131. [Google Scholar] [CrossRef] [PubMed]
- Alterio, V.; Hilvo, M.; di Fiore, A.; Supuran, C.T.; Pan, P.; Parkkila, S.; Scaloni, A.; Pastorek, J.; Pastorekova, S.; Pedone, C.; et al. Crystal structure of the catalytic domain of the tumor-associated human carbonic anhydrase IX. Proc. Natl. Acad. Sci. USA 2009, 106, 16233–16238. [Google Scholar] [CrossRef] [PubMed]
- Stams, T.; Nair, S.K.; Okuyama, T.; Waheed, A.; Sly, W.S.; Christianson, D.W. Crystal structure of the secretory form of membrane-associated human carbonic anhydrase IV at 2.8 Å resolution. Proc. Natl. Acad. Sci. USA 1996, 93, 13589–13594. [Google Scholar] [CrossRef] [PubMed]
- Whittington, D.A.; Waheed, A.; Ulmasov, B.; Shah, G.N.; Grubb, J.H.; Sly, W.S.; Christianson, D.W. Crystal structure of the dimeric extracellular domain of human carbonic anhydrase XII, a bitopic membrane protein overexpressed in certain cancer tumor cells. Proc. Natl. Acad. Sci. USA 2001, 98, 9545–9550. [Google Scholar] [CrossRef] [PubMed]
- Whittington, D.A.; Grubb, J.H.; Waheed, A.; Shah, G.N.; Sly, W.S.; Christianson, D.W. Expression, assay, and structure of the extracellular domain of murine carbonic anhydrase XIV: Implications for selective inhibition of membrane-associated isozymes. J. Biol. Chem. 2004, 279, 7223–7228. [Google Scholar] [CrossRef] [PubMed]
- Pilka, E.S.; Kochan, G.; Oppermann, U.; Yue, W.W. Crystal structure of the secretory isozyme of mammalian carbonic anhydrases CA VI: Implications for biological assembly and inhibitor development. Biochem. Biophys. Res. Commun. 2012, 419, 485–489. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, K.; Masuda, T.; Ohashi, H.; Mihara, H.; Suzuki, Y. Multiple proline substitutions cumulatively thermostabilize Bacillus cereus ATCC7064 oligo-1,6-glucosidase. Irrefragable proof supporting the proline rule. Eur. J. Biochem. 1994, 226, 277–283. [Google Scholar] [CrossRef] [PubMed]
- Vieille, C.; Zeikus, G.J. Hyperthermophilic enzymes: Sources, uses, and molecular mechanisms for thermostability. Microbiol. Mol. Biol. Rev. 2001, 65, 1–43. [Google Scholar] [CrossRef] [PubMed]
- Villbrandt, B.; Sagner, G.; Schomburg, D. Investigations on the thermostability and function of truncated Thermus aquaticus DNA polymerase fragments. Protein Eng. 1997, 10, 1281–1288. [Google Scholar] [CrossRef] [PubMed]
- Boone, C.D.; Rasi, V.; Tu, C.; McKenna, R. Structural and catalytic effects of proline substitution and surface loop deletion in the extended active site of human carbonic anhydrase II. FEBS J. 2015, 282, 1445–1457. [Google Scholar] [CrossRef] [PubMed]
- Kim, I.; Svendsen, H.F. Comparative study of the heats of absorption of post-combustion CO2 absorbents. Int. J. Greenh. Gas Control 2011, 5, 390–395. [Google Scholar] [CrossRef]
- Fox, R.J.; Davis, S.C.; Mundorff, E.C.; Newman, L.M.; Gavrilovic, V.; Ma, S.K.; Chung, L.M.; Ching, C.; Tam, S.; Muley, S.; et al. Improving catalytic function by ProSAR-driven enzyme evolution. Nat. Biotechnol. 2007, 25, 338–344. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Di Fiore, A.; Alterio, V.; Monti, S.M.; De Simone, G.; D'Ambrosio, K. Thermostable Carbonic Anhydrases in Biotechnological Applications. Int. J. Mol. Sci. 2015, 16, 15456-15480. https://doi.org/10.3390/ijms160715456
Di Fiore A, Alterio V, Monti SM, De Simone G, D'Ambrosio K. Thermostable Carbonic Anhydrases in Biotechnological Applications. International Journal of Molecular Sciences. 2015; 16(7):15456-15480. https://doi.org/10.3390/ijms160715456
Chicago/Turabian StyleDi Fiore, Anna, Vincenzo Alterio, Simona M. Monti, Giuseppina De Simone, and Katia D'Ambrosio. 2015. "Thermostable Carbonic Anhydrases in Biotechnological Applications" International Journal of Molecular Sciences 16, no. 7: 15456-15480. https://doi.org/10.3390/ijms160715456
APA StyleDi Fiore, A., Alterio, V., Monti, S. M., De Simone, G., & D'Ambrosio, K. (2015). Thermostable Carbonic Anhydrases in Biotechnological Applications. International Journal of Molecular Sciences, 16(7), 15456-15480. https://doi.org/10.3390/ijms160715456