Colloidal Stability & Conformational Changes in β-Lactoglobulin: Unfolding to Self-Assembly
Abstract
:1. Introduction
2. Results and Discussion
2.1. Evaluating the Colloidal Stability of βlg

| pH | Tagg (°C) a | kD (L/mg) at 25 °C | Z-Average (nm) |
|---|---|---|---|
| 5.5 | 65 | −4.13 | 6.94 |
| 7.0 | 56 | 0.28 | 6.87 |
| 8.5 | 52 | 2.24 | 3.63 |
2.2. Evaluating the Conformational Stability of βlg
2.2.1. Differential Scanning Calorimetry (DSC)

| pH | Tm (°C) a | ΔHv (kJ/mol) a | ΔHcal (kJ/mol) a | ΔHv a:ΔHcal a | Tm (°C) b | Tm (°C) c | ΔHv (kJ/mol) d | ΔHv (kJ/mol) e |
|---|---|---|---|---|---|---|---|---|
| 5.5 | 76.7 | 711.3 | 298.9 | 2.380 | 70.5 | 72.1 | 520 | 757 |
| 7.0 | 69.2 | 281.8 | 316.6 | 0.8901 | 62.9 | 66.0 | 364 | 434 |
| 8.5 | 66.0 | 250.1 | 232.0 | 1.078 | 59.1 | 62.0 | 171 | 250 |
2.2.2. Raman Spectroscopy



2.3. Mechanism of Unfolding and Aggregation
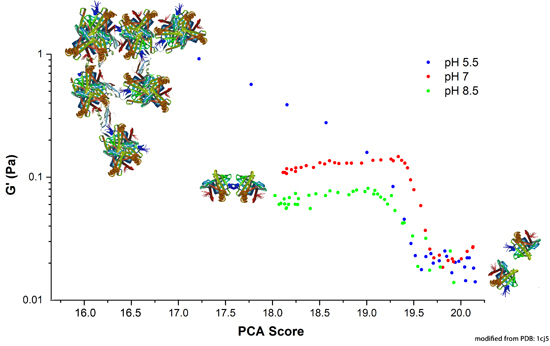
2.3.1. Correlation between Protein Structural Changes and Microstructure/Rheology


2.3.2. Raman Spectroscopy as a Means for Studying the Relative Interactions of Protein Unfolding and Aggregation

3. Experimental Section
4. Conclusions
Author Contributions
Conflicts of Interest
Appendixes

References
- Qiu, C.; Kivipelto, M.; von Strauss, E. Epidemiology of Alzheimer’s disease: Occurrence, determinants, and strategies toward intervention. Dialogues Clin. Neurosci. 2009, 11, 111. [Google Scholar] [PubMed]
- Chang, B.S.; Hershenson, S. Practical approaches to protein formulation development. Ration. Des. Stable Protein Formul. 2002, 13, 1–25. [Google Scholar]
- Amin, S.; Barnett, G.V.; Pathak, J.A.; Roberts, C.J.; Sarangapani, P.S. Protein aggregation, particle formation, characterization & rheology. Curr. Opin. Colloid Interface Sci. 2014, 19, 438–449. [Google Scholar]
- Mezzenga, R.; Fischer, P. The self-assembly, aggregation and phase transitions of food protein systems in one, two and three dimensions. Rep. Prog. Phys. 2013, 76, 046601. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Roberts, C.J. Aggregation of Therapeutic Proteins; John Wiley & Sons, Inc.: Hoboken, NJ, USA, 2012. [Google Scholar]
- Saluja, A.; Fesinmeyer, R.M.; Hogan, S.; Brems, D.N.; Gokarn, Y.R. Diffusion and sedimentation interaction parameters for measuring the second virial coefficient and their utility as predictors of protein aggregation. Biophys. J. 2010, 99, 2657–2665. [Google Scholar] [CrossRef] [PubMed]
- Connolly, B.D.; Petry, C.; Yadav, S.; Demeule, B.; Ciaccio, N.; Moore, J.M.; Shire, S.J.; Gokarn, Y.R. Weak interactions govern the viscosity of concentrated antibody solutions: High-throughput analysis using the diffusion interaction parameter. Biophys. J. 2012, 103, 69–78. [Google Scholar] [CrossRef] [PubMed]
- Xavier, K.A.; Willson, R.C. Association and dissociation kinetics of anti-hen egg lysozyme monoclonal antibodies hyhel-5 and hyhel-10. Biophys. J. 1998, 74, 2036–2045. [Google Scholar] [CrossRef]
- Sakurai, K.; Goto, Y. Manipulating monomer-dimer equilibrium of bovine β-Lactoglobulin by amino acid substitution. J. Biol. Chem. 2002, 277, 25735–25740. [Google Scholar] [CrossRef] [PubMed]
- Elofsson, U.M.; Dejmek, P.; Paulsson, M.A. Heat-induced aggregation of β-Lactoglobulin studied by dynamic light scattering. Int. Dairy J. 1996, 6, 343–357. [Google Scholar] [CrossRef]
- Haque, Z.U.; Sharma, M. Thermal gelation of β-Lactoglobulin ab purified from cheddar whey. 1. Effect of pH on association as observed by dynamic light scattering. J. Agric. Food Chem. 1997, 45, 2958–2963. [Google Scholar] [CrossRef]
- Phan-Xuan, T.; Durand, D.; Nicolai, T.; Donato, L.; Schmitt, C.; Bovetto, L. Heat induced formation of β-Lactoglobulin microgels driven by addition of calcium ions. Food Hydrocoll. 2014, 34, 227–235. [Google Scholar] [CrossRef]
- Verheul, M.; Roefs, S.P.; de Kruif, K.G. Kinetics of heat-induced aggregation of β-Lactoglobulin. J. Agric. Food Chem. 1998, 46, 896–903. [Google Scholar] [CrossRef]
- Vetri, V.; Militello, V. Thermal induced conformational changes involved in the aggregation pathways of β-Lactoglobulin. Biophys. Chem. 2005, 113, 83–91. [Google Scholar] [CrossRef] [PubMed]
- Aymard, P.; Durand, D.; Nicolai, T. The effect of temperature and ionic strength on the dimerisation of β-Lactoglobulin. Int. J. Biol. Macromol. 1996, 19, 213–221. [Google Scholar] [CrossRef]
- Baussay, K.; le Bon, C.; Nicolai, T.; Durand, D.; Busnel, J.-P. Influence of the ionic strength on the heat-induced aggregation of the globular protein β-Lactoglobulin at pH 7. Int. J. Biol. Macromol. 2004, 34, 21–28. [Google Scholar] [CrossRef] [PubMed]
- Majhi, P.R.; Ganta, R.R.; Vanam, R.P.; Seyrek, E.; Giger, K.; Dubin, P.L. Electrostatically driven protein aggregation: β-Lactoglobulin at low ionic strength. Langmuir 2006, 22, 9150–9159. [Google Scholar] [CrossRef] [PubMed]
- Schokker, E.; Singh, H.; Pinder, D.; Creamer, L. Heat-induced aggregation of β-Lactoglobulin ab at pH 2.5 as influenced by ionic strength and protein concentration. Int. Dairy J. 2000, 10, 233–240. [Google Scholar] [CrossRef]
- Jones, O.G.; Adamcik, J.; Handschin, S.; Bolisetty, S.; Mezzenga, R. Fibrillation of β-Lactoglobulin at low pH in the presence of a complexing anionic polysaccharide. Langmuir 2010, 26, 17449–17458. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.-M.; Savin, G.; Pouzot, M.; Schmitt, C.; Mezzenga, R. Structure of heat-induced β-Lactoglobulin aggregates and their complexes with sodium-dodecyl sulfate. Biomacromolecules 2008, 9, 2477–2486. [Google Scholar] [CrossRef] [PubMed]
- Van der Kooij, H.M.; Spruijt, E.; Voets, I.K.; Fokkink, R.; Cohen Stuart, M.A.; van der Gucht, J. On the stability and morphology of complex coacervate core micelles: From spherical to wormlike micelles. Langmuir 2012, 28, 14180–14191. [Google Scholar] [CrossRef] [PubMed]
- Dave, A.C.; Loveday, S.M.; Anema, S.G.; Jameson, G.B.; Singh, H. Modulating β-Lactoglobulin nanofibril self-assembly at pH 2 using glycerol and sorbitol. Biomacromolecules 2013, 15, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Gosal, W.S.; Clark, A.H.; Ross-Murphy, S.B. Fibrillar β-Lactoglobulin gels: Part 1. Fibril formation and structure. Biomacromolecules 2004, 5, 2408–2419. [Google Scholar] [CrossRef] [PubMed]
- Gosal, W.S.; Clark, A.H.; Ross-Murphy, S.B. Fibrillar β-Lactoglobulin gels: Part 2. Dynamic mechanical characterization of heat-set systems. Biomacromolecules 2004, 5, 2420–2429. [Google Scholar] [CrossRef] [PubMed]
- Gosal, W.S.; Clark, A.H.; Ross-Murphy, S.B. Fibrillar β-Lactoglobulin gels: Part 3. Dynamic mechanical characterization of solvent-induced systems. Biomacromolecules 2004, 5, 2430–2438. [Google Scholar] [CrossRef] [PubMed]
- Dave, A.C.; Loveday, S.M.; Anema, S.G.; Jameson, G.B.; Singh, H. Formation of nano-fibrils from the a, b and c variants of bovine β-Lactoglobulin. Int. Dairy J. 2015, 41, 64–67. [Google Scholar] [CrossRef]
- Adamcik, J.; Jung, J.-M.; Flakowski, J.; de Los Rios, P.; Dietler, G.; Mezzenga, R. Understanding amyloid aggregation by statistical analysis of atomic force microscopy images. Nat. Nanotechnol. 2010, 5, 423–428. [Google Scholar] [CrossRef] [PubMed]
- Bolisetty, S.; Adamcik, J.; Mezzenga, R. Snapshots of fibrillation and aggregation kinetics in multistranded amyloid β-Lactoglobulin fibrils. Soft Matter 2011, 7, 493–499. [Google Scholar] [CrossRef]
- Lara, C.; Adamcik, J.; Jordens, S.; Mezzenga, R. General self-assembly mechanism converting hydrolyzed globular proteins into giant multistranded amyloid ribbons. Biomacromolecules 2011, 12, 1868–1875. [Google Scholar] [CrossRef] [PubMed]
- Lewis, E.N.; Qi, W.; Kidder, L.H.; Amin, S.; Kenyon, S.M.; Blake, S. Combined dynamic light scattering and Raman spectroscopy approach for characterizing the aggregation of therapeutic proteins. Molecules 2014, 19, 20888–20905. [Google Scholar] [CrossRef] [PubMed]
- Zhou, C.; Qi, W.; Lewis, E.N.; Carpenter, J.F. Concomitant Raman spectroscopy and dynamic light scattering for characterization of therapeutic proteins at high concentrations. Anal. Biochem. 2015, 472, 7–20. [Google Scholar] [CrossRef] [PubMed]
- Roberts, D.; Keeling, R.; Tracka, M.; van der Walle, C.; Uddin, S.; Warwicker, J.; Curtis, R. The role of electrostatics in protein–protein interactions of a monoclonal antibody. Mol. Pharm. 2014, 11, 2475–2489. [Google Scholar] [CrossRef] [PubMed]
- Casal, H.L.; Köhler, U.; Mantsch, H.H. Structural and conformational changes of β-Lactoglobulin B: An infrared spectroscopic study of the effect of pH and temperature. Biochim. Biophys. Acta 1988, 957, 11–20. [Google Scholar] [CrossRef]
- Harding, S.E.C.; Babur, Z. Protein-ligand Interactions: Hydrodynamics and Calorimetry (Practical Approach Series); Oxford University Press: Oxford, UK, 2001; p. 354. [Google Scholar]
- Gotham, S.M.; Fryer, P.J.; Pritchard, A.M. β-Lactoglobulin denaturation and aggregation reactions and fouling deposit formation: A DSC study. Int. J. Food Sci. Technol. 1992, 27, 313–327. [Google Scholar] [CrossRef]
- Ngarize, S.; Herman, H.; Adams, A.; Howell, N. Comparison of changes in the secondary structure of unheated, heated, and high-pressure-treated β-Lactoglobulin and ovalbumin proteins using fourier transform Raman spectroscopy and self-deconvolution. J. Agric. Food Chem. 2004, 52, 6470–6477. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.Q. Raman spectroscopy of protein pharmaceuticals. J. Pharm. Sci. 2007, 96, 2861–2878. [Google Scholar] [CrossRef] [PubMed]
- Ludwig, M.; Asher, S.A. Ultraviolet resonance Raman excitation profiles of tyrosine: Dependence of Raman cross sections on excited-state intermediates. J. Am. Chem. Soc. 1988, 110, 1005–1011. [Google Scholar] [CrossRef]
- Amin, S.; Blake, S.; Kenyon, S.; Kennel, R.; Lewis, E. A novel combination of DLS-optical microrheology and low frequency Raman spectroscopy to reveal underlying biopolymer self-assembly and gelation mechanisms. J. Chem. Phys. 2014, 141, 234201. [Google Scholar] [CrossRef] [PubMed]
- Amin, S.; Blake, S.; Kennel, R.C.; Lewis, E.N. Revealing new structural insights from surfactant micelles through DLS, microrheology and Raman spectroscopy. Materials 2015, 8, 3754–3766. [Google Scholar] [CrossRef]
- Worldwide Protein Data Bank. Available online: http://www.rcsb.org/pdb/home/home.do (accessed on 30 July 2015).
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Blake, S.; Amin, S.; Qi, W.; Majumdar, M.; Lewis, E.N. Colloidal Stability & Conformational Changes in β-Lactoglobulin: Unfolding to Self-Assembly. Int. J. Mol. Sci. 2015, 16, 17719-17733. https://doi.org/10.3390/ijms160817719
Blake S, Amin S, Qi W, Majumdar M, Lewis EN. Colloidal Stability & Conformational Changes in β-Lactoglobulin: Unfolding to Self-Assembly. International Journal of Molecular Sciences. 2015; 16(8):17719-17733. https://doi.org/10.3390/ijms160817719
Chicago/Turabian StyleBlake, Steven, Samiul Amin, Wei Qi, Madhabi Majumdar, and E. Neil Lewis. 2015. "Colloidal Stability & Conformational Changes in β-Lactoglobulin: Unfolding to Self-Assembly" International Journal of Molecular Sciences 16, no. 8: 17719-17733. https://doi.org/10.3390/ijms160817719
APA StyleBlake, S., Amin, S., Qi, W., Majumdar, M., & Lewis, E. N. (2015). Colloidal Stability & Conformational Changes in β-Lactoglobulin: Unfolding to Self-Assembly. International Journal of Molecular Sciences, 16(8), 17719-17733. https://doi.org/10.3390/ijms160817719