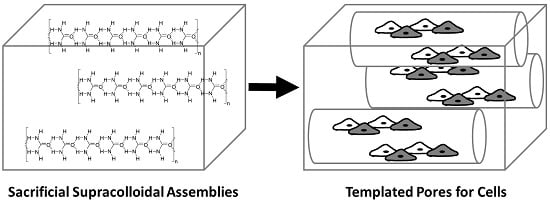
Supracolloidal Assemblies as Sacrificial Templates for Porous Silk-Based Biomaterials
Abstract
:1. Introduction

2. Results and Discussion
2.1. Urea Solubility in Non-Aqueous Solvents
| Solvent | ε | ET | π* | α | Β | Boiling Point (°C) | Surface Tension (mN/m) | Solubility of Urea |
|---|---|---|---|---|---|---|---|---|
| Cyclohexane | 2.10 | 0.006 | 0.00 | 0.00 | 0.00 | 80.7 | 25.0 | I.S. |
| Toluene | 2.38 | 0.099 | 0.49 | 0.00 | 0.11 | 110.6 | 28.5 | I.S. |
| Chloroform | 4.80 | 0.259 | 0.69 | 0.44 | 0.00 | 61.2 | 26.7 | I.S. |
| Tetrahydrofuran | 7.58 | 0.207 | 0.55 | 0.00 | 0.55 | 66.0 | 26.4 | I.S. |
| Dichloromethane | 8.93 | 0.309 | 0.73 | 0.30 | 0.00 | 39.6 | 26.5 | I.S. |
| Ethyl acetate | 36.6 | 0.18 | 0.55 | 0.00 | 0.45 | 77.1 | 23.8 | I.S. |
| Acetonitrile | 45.60 | 0.460 | 0.75 | 0.19 | 0.40 | 82.0 | 19.1 | I.S. |
| Isopropanol | 49.20 | 0.570 | 0.48 | 0.76 | 0.84 | 82.6 | 23.0 | I.S. |
| Butanol | 50.20 | 0.600 | 0.47 | 0.84 | 0.84 | 117.4 | 24.2 | I.S. |
| Ethanol | 51.90 | 0.650 | 0.54 | 0.86 | 0.75 | 78.4 | 22.3 | I.S. |
| Methanol | 55.40 | 0.760 | 0.60 | 0.98 | 0.66 | 64.7 | 22.5 | S |
| Formic acid | 57.70 | 0.830 | 0.65 | 1.23 | 0.38 | 100.8 | 37.7 | S |
| Hexafluoroisopropanol | 65.30 | 1.070 | 0.65 | 1.96 | 0.00 | 58.2 | 16.1 | S |
| Hexafluoroacetone∙3H2O | N.R. | N.R. | N.R. | N.R. | N.R. | −26.0 a | N.R. | S |
| Water | 63.10 | 1.000 | 1.09 | 1.17 | 0.47 | 100.0 | 72.8 | S |
2.2. The Role of Solvent Choice on the Morphology of the Sacrificial Supracolloidal Porogens

2.3. Supracolloidal Templation of Porous Silk Biomaterials


3. Experimental Section
3.1. Materials
3.2. Urea Crystallization from Non-Aqueous Solvents
3.3. Urea Crystal Templating of Porous Silk-Based Films
3.4. Preparation of Silk-Based Tissue Scaffolds with Aligned Pores
3.5. Optical Microscopy of Urea and Silk Materials
3.6. Scanning Electron Microscopy (SEM)
3.7. In Vitro Cell Culture
4. Conclusions
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Lutolf, M.P.; Hubbell, J.A. Synthetic biomaterials as instructive extracellular microenvironments for morphogenesis in tissue engineering. Nat. Biotechnol. 2005, 23, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Edalat, F.; Sheu, I.; Manoucheri, S.; Khademhosseini, A. Material strategies for creating artificial cell-instructive niches. Curr. Opin. Biotechnol. 2012, 23, 820–825. [Google Scholar] [CrossRef] [PubMed]
- Rice, J.J.; Martino, M.M.; de Laporte, L.; Tortelli, F.; Briquez, P.S.; Hubbell, J.A. Engineering the regenerative microenvironment with biomaterials. Adv. Healthc. Mater. 2013, 2, 57–71. [Google Scholar] [CrossRef] [PubMed]
- Custódio, C.A.; Reis, R.L.; Mano, J.F. Engineering biomolecular microenvironments for cell instructive biomaterials. Adv. Healthc. Mater. 2014, 3, 797–810. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rockwood, D.N.; Preda, R.C.; Yücel, T.; Wang, X.; Lovett, M.L.; Kaplan, D.L. Materials fabrication from Bombyx mori silk fibroin. Nat. Protoc. 2011, 6, 1612–1631. [Google Scholar] [CrossRef] [PubMed]
- Torres-Rendon, J.G.; Femmer, T.; de Laporte, L.; Tigges, T.; Rahimi, K.; Gremse, F.; Zafarnia, S.; Lederle, W.; Ifuku, S.; Wessling, M.; et al. Bioactive gyroid scaffolds formed by sacrificial templating of nanocellulose and nanochitin hydrogels as instructive platforms for biomimetic tissue engineering. Adv. Mater. 2015, 27, 2989–2995. [Google Scholar] [CrossRef] [PubMed]
- Sutherland, T.D.; Young, J.H.; Weisman, S.; Hayashi, C.Y.; Merritt, D.J. Insect silk: One name, many materials. Annu. Rev. Entomol. 2010, 55, 171–188. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.G.; Römer, L.M.; Scheibel, T.R. Polymeric materials based on silk proteins. Polymer 2008, 49, 4309–4327. [Google Scholar] [CrossRef]
- Kundu, B.; Rajkhowa, R.; Kundu, S.C.; Wang, X. Silk fibroin biomaterials for tissue regenerations. Adv. Drug Deliv. Rev. 2013, 65, 457–470. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.G.; Scheibel, T.R. Composite materials based on silk proteins. Prog. Polym. Sci. 2010, 35, 1093–1115. [Google Scholar] [CrossRef]
- Preda, R.C.; Leisk, G.; Omenetto, F.; Kaplan, D.L. Bioengineered silk proteins to control cell and tissue functions. Methods Mol. Biol. 2013, 996, 19–41. [Google Scholar] [PubMed]
- Hardy, J.G.; Scheibel, T.R. Production and processing of spider silk proteins. J. Polym. Sci. A Polym. Chem. 2009, 47, 3957–3963. [Google Scholar] [CrossRef]
- Schacht, K.; Scheibel, T. Processing of recombinant spider silk proteins into tailor-made materials for biomaterials applications. Curr. Opin. Biotechnol. 2014, 29, 62–69. [Google Scholar] [CrossRef] [PubMed]
- Widhe, M.; Johansson, J.; Hedhammar, M.; Rising, A. Invited review current progress and limitations of spider silk for biomedical applications. Biopolymers 2012, 97, 468–478. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.G.; Scheibel, T.R. Silk-inspired polymers and proteins. Biochem. Soc. Trans. 2009, 37, 677–681. [Google Scholar] [CrossRef] [PubMed]
- Moffat, J.R.; Seeley, G.J.; Carter, J.T.; Burgess, A.; Smith, D.K. Nanostructured polymers with embedded self-assembled reactive gel networks. Chem. Commun. 2008, 4601–4603. [Google Scholar] [CrossRef] [PubMed]
- Zawko, S.A.; Schmidt, C.E. Crystal templating dendritic pore networks and fibrillar microstructure into hydrogels. Acta Biomater. 2010, 6, 2415–2421. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.G.; Cornelison, R.C.; Sukhavasi, R.C.; Saballos, R.J.; Vu, P.; Kaplan, D.L.; Schmidt, C.E. Electroactive tissue scaffolds with aligned pores as instructive platforms for biomimetic tissue engineering. Bioengineering 2015, 2, 15–34. [Google Scholar] [CrossRef]
- Lan, Y.; Corradini, M.G.; Liu, X.; May, T.E.; Borondics, F.; Weiss, R.G.; Rogers, M.A. Comparing and correlating solubility parameters governing the self-assembly of molecular gels using 1,3:2,4-dibenzylidene sorbitol as the gelator. Langmuir 2014, 30, 14128–14142. [Google Scholar] [CrossRef] [PubMed]
- Lan, L.; Corradini, M.G.; Rogers, M.A. Do molecular gelators cluster in hansen space? Cryst. Growth Des. 2014, 14, 4811–4818. [Google Scholar] [CrossRef]
- Hirst, A.R.; Smith, D.K. Solvent effects on supramolecular gel-phase materials: Two-component dendritic gel. Langmuir 2004, 20, 10851–10857. [Google Scholar] [CrossRef] [PubMed]
- Hardy, J.G.; Hirst, A.R.; Ashworth, I.; Brennan, C.; Smith, D.K. Exploring molecular recognition pathways within a family of gelators with different hydrogen bonding motifs. Tetrahedron 2007, 63, 7397–7406. [Google Scholar] [CrossRef]
- Edwards, W.; Lagadec, C.A.; Smith, D.K. Solvent-gelator interactions-using empirical solvent parameters to better understand the self-assembly of gel-phase materials. Soft Matter 2011, 7, 110–117. [Google Scholar] [CrossRef]
- Hardy, J.G.; Hirst, A.R.; Smith, D.K. Exploring molecular recognition pathways in one- and two-component gels formed by dendritic lysine-based gelators. Soft Matter 2012, 8, 3399–3406. [Google Scholar] [CrossRef]
- Rajan, R.; Awasthi, S.K.; Bhattacharjya, S.; Balaram, P. “Teflon-coated peptides”: Hexafluoroacetone trihydrate as a structure stabilizer for peptides. Biopolymers 1997, 42, 125–128. [Google Scholar] [CrossRef]
- Aldaye, F.A.; Palmer, A.L.; Sleiman, H.F. Assembling materials with DNA as the guide. Science 2008, 321, 1795–1799. [Google Scholar] [CrossRef] [PubMed]
- Carneiro, K.M.; Avakyan, N.; Sleiman, H.F. Long-range assembly of DNA into nanofibers and highly ordered networks. Wiley Interdiscip. Rev. Nanomed. Nanobiotechnol. 2013, 5, 266–285. [Google Scholar] [CrossRef] [PubMed]
- McLaughlin, C.K.; Hamblin, G.D.; Sleiman, H.F. Supramolecular DNA assembly. Chem. Soc. Rev. 2011, 40, 5647–5656. [Google Scholar] [CrossRef] [PubMed]
- Lin, N.; Liu, X.Y. Correlation between hierarchical structure of crystal networks and macroscopic performance of mesoscopic soft materials and engineering principles. Chem. Soc. Rev. 2015. [Google Scholar] [CrossRef]
- Lo, P.K.; Metera, K.L.; Sleiman, H.F. Self-assembly of three-dimensional DNA nanostructures and potential biological applications. Curr. Opin. Chem. Biol. 2010, 14, 597–607. [Google Scholar] [CrossRef] [PubMed]
- Stulz, E. DNA architectonics: Towards the next generation of bio-inspired materials. Chemistry 2012, 18, 4456–4469. [Google Scholar] [CrossRef] [PubMed]
- Jaeger, L.; Chworos, A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 2006, 16, 531–543. [Google Scholar] [CrossRef] [PubMed]
- Jacobs, W.M.; Oxtoby, D.W.; Frenkel, D. Phase separation in solutions with specific and nonspecific interactions. J. Chem. Phys. 2014, 140, 204109. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jacobs, W.M.; Reinhardt, A.; Frenkel, D. Rational design of self-assembly pathways for complex multicomponent structures. PNAS 2015, 112, 6313–6318. [Google Scholar] [CrossRef] [PubMed]
- Frenkel, D. Why colloidal systems can be described by statistical mechanics: some not very original comments on the Gibbs paradox. Mol. Phys. 2014, 112, 2325–2329. [Google Scholar] [CrossRef]
- Ciesielski, A.; Palma, C.A.; Bonini, M.; Samorì, P. Towards supramolecular engineering of functional nanomaterials: Pre-programming multi-component 2D self-assembly at solid-liquid interfaces. Adv. Mater. 2010, 22, 3506–3520. [Google Scholar] [CrossRef] [PubMed]
- Guo, R.; Mao, J.; Xie, X.M.; Yan, L.T. Predictive supracolloidal helices from patchy particles. Sci. Rep. 2014, 4, 7021. [Google Scholar] [CrossRef] [PubMed]
© 2015 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hardy, J.G.; Ghezzi, C.E.; Saballos, R.J.; Kaplan, D.L.; Schmidt, C.E. Supracolloidal Assemblies as Sacrificial Templates for Porous Silk-Based Biomaterials. Int. J. Mol. Sci. 2015, 16, 20511-20522. https://doi.org/10.3390/ijms160920511
Hardy JG, Ghezzi CE, Saballos RJ, Kaplan DL, Schmidt CE. Supracolloidal Assemblies as Sacrificial Templates for Porous Silk-Based Biomaterials. International Journal of Molecular Sciences. 2015; 16(9):20511-20522. https://doi.org/10.3390/ijms160920511
Chicago/Turabian StyleHardy, John G., Chiara E. Ghezzi, Richard J. Saballos, David L. Kaplan, and Christine E. Schmidt. 2015. "Supracolloidal Assemblies as Sacrificial Templates for Porous Silk-Based Biomaterials" International Journal of Molecular Sciences 16, no. 9: 20511-20522. https://doi.org/10.3390/ijms160920511
APA StyleHardy, J. G., Ghezzi, C. E., Saballos, R. J., Kaplan, D. L., & Schmidt, C. E. (2015). Supracolloidal Assemblies as Sacrificial Templates for Porous Silk-Based Biomaterials. International Journal of Molecular Sciences, 16(9), 20511-20522. https://doi.org/10.3390/ijms160920511