Over-Expression of GmGIa-Regulated Soybean miR172a Confers Early Flowering in Transgenic Arabidopsis thaliana
Abstract
:1. Introduction
2. Results
2.1. Identification and Analysis of gma-miR172a and Glyma03g33470 Sequences
2.2. Temporal and Spatial Expression Patterns of gma-miR172 and Their Target Genes in Soybean
2.3. Diurnal Rhythm of gma-miR172a and Glyma03g33470
2.4. Over-Expression of gma-miR172a Results in Earlier Flowering in Transgenic Arabidopsis
2.5. Up-Regulation of FT, AP1 and LFY in gma-miR172a-Transgenic Plants
2.6. toe1 Mutant Plants Restores Earlier Flowering Phenotype by the Expression of Glyma03g33470
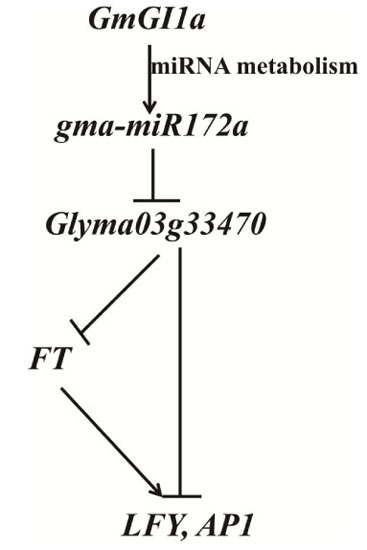
2.7. GmGIa Regulates gma-miR172a through miRNA Metabolism in Soybean
3. Discussion
4. Materials and Methods
4.1. Plant Materials and Growth Conditions
4.2. Real Time RT-PCR Analyses
4.3. RACE Mapping of miRNA Target Cleavage Sites
4.4. Gene Constructs and Generation of Transgenic Arabidopsis Plants
4.5. Flowering Time Measurements
4.6. Statistical Analysis
Supplementary Materials
Acknowledgments
Author Contributions
Conflicts of Interest
References
- Huijser, P.; Schmid, M. The control of developmental phase transitions in plants. Development 2011, 138, 4117–4129. [Google Scholar] [CrossRef] [PubMed]
- Mouradov, A.; Cremer, F.; Coupland, G. Control of flowering time: Interacting pathways as a basis for diversity. Plant Cell 2002, 14, 111–130. [Google Scholar]
- Yamaguchi, A.; Abe, M. Regulation of reproductive development by non-coding RNA in Arabidopsis: To flower or not to flower. J. Plant Res. 2012, 125, 693–704. [Google Scholar] [CrossRef] [PubMed]
- Fan, T.; Li, X.; Yang, W.; Xia, K.; Ouyang, J.; Zhang, M. Rice osa-miR171c mediates phase change from vegetative to reproductive development and shoot apical meristem maintenance by repressing four OsHAM transcription factors. PLoS ONE 2015, 10, e0125833. [Google Scholar] [CrossRef] [PubMed]
- Adams, S.; Allen, T.; Whitelam, G.C. Interaction between the light quality and flowering time pathways in Arabidopsis. Plant J. 2009, 60, 257–267. [Google Scholar] [CrossRef] [PubMed]
- Schiessl, S.; Iniguez-Luy, F.; Qian, W.; Snowdon, R.J. Diverse regulatory factors associate with flowering time and yield responses in winter-type Brassica napus. BMC Genom. 2015, 16. [Google Scholar] [CrossRef] [PubMed]
- Shim, J.S.; Imaizumi, T. Circadian clock and photoperiodic response in Arabidopsis: From seasonal flowering to redox homeostasis. Biochemistry 2015, 54, 157–170. [Google Scholar] [CrossRef] [PubMed]
- Jones-Rhoades, M.; Bartel, D.P.; Bartel, B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006, 57, 19–53. [Google Scholar] [CrossRef] [PubMed]
- Kim, W.; Ahn, H.J.; Chiou, T.J.; Ahn, J.H. The role of the miR399-PHO2 module in the regulation of flowering time in response to different ambient temperatures in Arabidopsis thaliana. Mol. Cells 2011, 32, 83–88. [Google Scholar] [CrossRef] [PubMed]
- Rubio-Somoza, I.; Weigel, D. MicroRNA networks and developmental plasticity in plants. Trends Plant Sci. 2011, 16, 258–264. [Google Scholar] [CrossRef] [PubMed]
- Jin, D.; Wang, Y.; Zhao, Y.; Chen, M. MicroRNAs and their crosstalks in plant development. J. Genet. Genom. 2013, 40, 161–170. [Google Scholar] [CrossRef] [PubMed]
- Schwab, R.; Palatnik, J.F.; Riester, M.; Schommer, C.; Schmid, M.; Weigel, D. Specific effects of microRNAs on the plant transcriptome. Dev. Cell 2005, 8, 517–527. [Google Scholar] [CrossRef] [PubMed]
- Wu, G.; Poethig, R.S. Temporal regulation of shoot development in Arabidopsis thaliana by miR156 and its target SPL3. Development 2006, 133, 3539–3547. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.J.; Lee, J.H.; Kim, W.; Jung, H.S.; Huijser, P.; Ahn, J.H. The microRNA156-SQUAMOSA PROMOTER BINDING PROTEIN-LIKE3 module regulates ambient temperature-responsive flowering via FLOWERING LOCUS T in Arabidopsis. Plant Physiol. 2012, 159, 461–478. [Google Scholar] [CrossRef] [PubMed]
- Achard, P.; Herr, A.; Baulcombe, D.C.; Harberd, N.P. Modulation of floral development by a gibberellin-regulated microRNA. Development 2004, 131, 3357–3365. [Google Scholar] [CrossRef] [PubMed]
- Park, W.; Li, J.; Song, R.; Messing, J.; Chen, X. CARPEL FACTORY, a Dicer homolog, and HEN1, a novel protein, act in microRNA metabolism in Arabidopsis thaliana. Curr. Biol. 2002, 3, 1484–1495. [Google Scholar] [CrossRef]
- Axtell, M.J.; Bartel, D.P. Antiquity of microRNAs and their targets in land plants. Plant Cell 2005, 17, 1658–1673. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.H.; Helliwell, C.A. Regulation of flowering time and floral patterning by miR172. J. Exp. Bot. 2011, 62, 487–495. [Google Scholar] [CrossRef] [PubMed]
- Aukerman, M.J.; Sakai, H. Regulation of flowering time and floral organ identity by a microRNA and its APETALA2-like target genes. Plant Cell 2003, 15, 2730–2741. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Wang, L.; Zeng, L.; Zhang, C.; Ma, H. Arabidopsis TOE proteins convey a photoperiodic signal to antagonize CONSTANS and regulate flowering time. Genes Dev. 2015, 29, 975–987. [Google Scholar] [CrossRef] [PubMed]
- Mathieu, J.; Yant, L.J.; Mürdter, F.; Küttner, F.; Schmid, M. Repression of flowering by the miR172 target SMZ. PLoS Biol. 2009, 7, e1000148. [Google Scholar] [CrossRef] [PubMed]
- Zhu, Q.H.; Upadhyaya, N.M.; Gubler, F.; Helliwell, C.A. Over-expression of miR172 causes loss of spikelet determinacy and floral organ abnormalities in rice (Oryza sativa). BMC Plant Biol. 2009, 9. [Google Scholar] [CrossRef] [PubMed]
- Wang, Y.; Wang, L.; Zou, Y.; Chen, L.; Cai, Z.; Zhang, S.; Zhao, F.; Tian, Y.; Jiang, Q.; Ferguson, B.J.; et al. Soybean miR172c targets the repressive AP2 transcription factor NNC1 to activate ENOD40 expression and regulate nodule initiation. Plant Cell 2014, 26, 4782–4801. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Cao, D.; Huang, Z.; Wang, J.; Lu, S.; Xu, Y.; Liu, B.; Kong, F.; Yuan, X. Dual functions of GmTOE4a in the regulation of photoperiod-mediated flowering and plant morphology in soybean. Plant Mol. Biol. 2015, 88, 343–355. [Google Scholar] [CrossRef] [PubMed]
- Jung, J.H.; Seo, Y.H.; Seo, P.J.; Reyes, J.L.; Yun, J.; Chua, N.H.; Park, C.M. The GIGANTEA-regulated microRNA172 mediates photoperiodic flowering independent of CONSTANS in Arabidopsis. Plant Cell 2007, 19, 2736–2748. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Zhang, X.; Hu, R.; Wu, F.; Ma, J.; Meng, Y.; Fu, Y. Identification and molecular characterization of FKF1 and GI homologous genes in soybean. PLoS ONE 2013, 8, e79036. [Google Scholar] [CrossRef] [PubMed]
- Watanabe, S.; Xia, Z.; Hideshima, R.; Tsubokura, Y.; Sato, S.; Yamanaka, N.; Takahashi, R.; Anai, T.; Tabata, S.; Kitamura, K.; et al. A map-based cloning strategy employing a residual heterozygous line reveals that the GIGANTEA gene is involved in soybean maturity and flowering. Genetics 2011, 188, 395–407. [Google Scholar] [CrossRef] [PubMed]
- Voinnet, O. Origin, biogenesis, and activity of plant microRNAs. Cell 2009, 136, 669–687. [Google Scholar] [CrossRef] [PubMed]
- Spanudakis, E.; Jackson, S. The role of microRNAs in the control of flowering time. J. Exp. Bot. 2014, 65, 365–380. [Google Scholar] [CrossRef] [PubMed]
- Srikanth, A.; Schmid, M. Regulation of flowering time: All roads lead to Rome. Cell. Mol. Life Sci. 2011, 68, 2013–2037. [Google Scholar] [CrossRef] [PubMed]
- Sun, L.M.; Ai, X.Y.; Li, W.Y.; Guo, W.W.; Deng, X.X.; Hu, C.G.; Zhang, J.Z. Identification and comparative profiling of miRNAs in an early flowering mutant of trifoliate orange and its wild type by genome-wide deep sequencing. PLoS ONE 2012, 7, e43760. [Google Scholar] [CrossRef] [PubMed]
- Chen, Z.X.; Li, F.L.; Yang, S.N.; Dong, Y.B.; Yuan, Q.H.; Wang, F.; Li, W.M.; Jiang, Y.; Jia, S.R.; Pei, X.W. Identification and functional analysis of flowering related microRNAs in common wild rice (Oryza rufipogon Griff.). PLoS ONE 2013, 8, e82844. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.J.; Huang, J.Q.; Huang, Y.J.; Li, Z.; Zheng, B.S. Discovery and profiling of novel and conserved microRNAs during flower development in Carya cathayensis via deep sequencing. Planta 2012, 236, 613–621. [Google Scholar] [CrossRef] [PubMed]
- Ao, Y.; Wang, Y.W.; Chen, W.; Wang, T.; Yu, H.Y.; Zhang, Z.X. Identification and comparative profiling of microRNAs in wild-type Xanthoceras sorbifolia and its double flower mutant. Genes Genom. 2012, 34, 561–568. [Google Scholar] [CrossRef]
- Song, Y.; Ma, K.; Ci, D.; Zhang, Z.; Zhang, D. Sexual dimorphism floral microRNA profiling and target gene expression in andromonoecious poplar. PLoS ONE 2013, 7, e43760. [Google Scholar] [CrossRef] [PubMed]
- Nie, S.; Xu, L.; Wang, Y.; Huang, D.; Muleke, E.M.; Sun, X.; Wang, R.; Xie, Y.; Gong, Y.; Liu, L. Identification of bolting-related microRNAs and their targets reveals complex miRNA-mediated flowering-time regulatory networks in radish (Raphanus sativus L.). Sci. Rep. 2015, 5. [Google Scholar] [CrossRef] [PubMed]
- Li, W.B.; Wang, P.P.; Li, Y.G.; Zhang, K.X.; Ding, F.Q.; Nie, T.K.; Yang, X.; Lv, Q.X.; Zhao, L. Identification of microRNAs in response to different day lengths in soybean using high-throughput sequencing and qRT-PCR. PLoS ONE 2015, 10, e0132621. [Google Scholar] [CrossRef] [PubMed]
- Chen, X. A microRNA as a translational repressor of APETALA2 in Arabidopsis flower development. Science 2004, 303, 2022–2025. [Google Scholar] [CrossRef] [PubMed]
- Livak, K.J.; Schmittgen, T.D. Analysis of relative gene expression data using real-time quantitative PCR and the the 2−ΔΔCt method. Methods 2001, 25, 402–408. [Google Scholar] [CrossRef] [PubMed]








| Locus | Mature Sequence |
|---|---|
| gma-miR172a/b | AGAAUCUUGAUGAUGCUGCAU |
| gma-miR172c | GGAAUCUUGAUGAUGCUGCAG |
| gma-miR172d/e | GGAAUCUUGAUGAUGCUGCAGCAG |
| gma-miR172f | AGAAUCUUGAUGAUGCUGCA |
| gma-miR172g | GCAGCACCAUCAAGAUUCAC |
| gma-miR172h/i/j | GCAGCAGCAUCAAGAUUCACA |
| gma-miR172k | UGAAUCUUGAUGAUGCUGCAU |
| gma-miR172l | GGAAUCUUGAUGAUGCUGCAU |
| Primer Names | Primer Sequences (5′–3′) |
|---|---|
| Adaptor primer 1 | 5′-CCATCCTAATACGACTCACTATAGGGC-3′ |
| Adaptor primer 2 | 5′-ACTCACTATAGGGCTCGAGCGGC-3′ |
| Outer primer | 5′-GTGGCGACTAACTAACGGAAGCAAAG-3′ |
| Inner primer | 5′-TCACAACTATGACCCACAA-3′ |
| overlapping PCR-F | 5′-GAAGATCTCCCAGCAACGGGAGTGGTATGA-3′ |
| overlapping PCR-Rm | 5′-ATCCAGTAGATGCTGCAGTAGAGAAGG-3′ |
| overlapping PCR-Fm | 5′-ATCTACTGGATTCCCTTCAACAATAAT-3′ |
| overlapping PCR-R | 5′-GGGTAACCGTGGCGACTAACTAACGGAAGCAAAG-3′ |
| qGmACTIN4-F | 5′-GTGTCAGCCATACTGTCCCCATTT-3′ |
| qGmACTIN4-R | 5′-GTTTCAAGCTCTTGCTCGTAATCA-3′ |
| AtACTIN8-F | 5′-CGTCCCTGCCCTTTGTACAC-3′ |
| AtACTIN8-R | 5′-CGAACACTTCACCGGATCATT-3′ |
| gma-miR172a-F | 5′-AGATCTGTGAAGTCGTTTATGGCTGAT-3′ |
| gma-miR172a-R | 5′-GGTAACCTTAACAGTCGTTATTTGCGG-3′ |
| qGmDCL1-F | 5′-AAATGCGGACCTACCAAA-3′ |
| qGmDCL1-R | 5′-TCAAAGCGAATAACGACA -3′ |
| qGmSE-F | 5′-CTCACTGGGTGGTTGGTT-3′ |
| qGmSE-R | 5′-GATACATCCCTCGGCTCA-3′ |
| qGlyma03g33470-F | 5′-GCTTCTCCGTAGCATCTGG-3′ |
| qGlyma03g33470-R | 5′-GTGGAGGAATGTCATGTTTG-3′ |
| AtFLC-F | 5′-GCTCTTCTCGTCGTCTCC-3′ |
| AtFLC-R | 5′-GTTCGGTCTTCTTGGCTC-3′ |
| AtCO-F | 5′-AAGGTGATAAGGATGCCAAGGAG-3′ |
| AtCO-R | 5′-GGAGCCATATTTGATATTGAACTGA-3′ |
| AtSOC1-F | 5′-TCAGAACTTGGGCTACTC-3′ |
| AtSOC1-R | 5′-TTCTCGTCGTCTCCGCCTCC-3′ |
| AtAP1-F | 5′-TAAGCACATCCGCACTAG-3′ |
| AtAP1-R | 5′-TTCTTGATACAGACCACCC-3′ |
| AtFT-F | 5′-TGGTGGAGAAGACCTCAGGAAC-3′ |
| AtFT-R | 5′-TGCCAAGCTGTCGAAACAATAT-3′ |
| AtLFY-F | 5′-TGTGAACATCGCTTGTCGTC-3′ |
| AtLFY-R | 5′-TAATACCGCCAACTAAAGCC-3′ |
© 2016 by the authors; licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC-BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Sun, M.-Y.; Wang, X.-S.; Li, W.-B.; Li, Y.-G. Over-Expression of GmGIa-Regulated Soybean miR172a Confers Early Flowering in Transgenic Arabidopsis thaliana. Int. J. Mol. Sci. 2016, 17, 645. https://doi.org/10.3390/ijms17050645
Wang T, Sun M-Y, Wang X-S, Li W-B, Li Y-G. Over-Expression of GmGIa-Regulated Soybean miR172a Confers Early Flowering in Transgenic Arabidopsis thaliana. International Journal of Molecular Sciences. 2016; 17(5):645. https://doi.org/10.3390/ijms17050645
Chicago/Turabian StyleWang, Tao, Ming-Yang Sun, Xue-Song Wang, Wen-Bin Li, and Yong-Guang Li. 2016. "Over-Expression of GmGIa-Regulated Soybean miR172a Confers Early Flowering in Transgenic Arabidopsis thaliana" International Journal of Molecular Sciences 17, no. 5: 645. https://doi.org/10.3390/ijms17050645
APA StyleWang, T., Sun, M. -Y., Wang, X. -S., Li, W. -B., & Li, Y. -G. (2016). Over-Expression of GmGIa-Regulated Soybean miR172a Confers Early Flowering in Transgenic Arabidopsis thaliana. International Journal of Molecular Sciences, 17(5), 645. https://doi.org/10.3390/ijms17050645